
Carrot red leaf luteovirus associated RNA
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 8.39
Get precalculated fractions of proteins
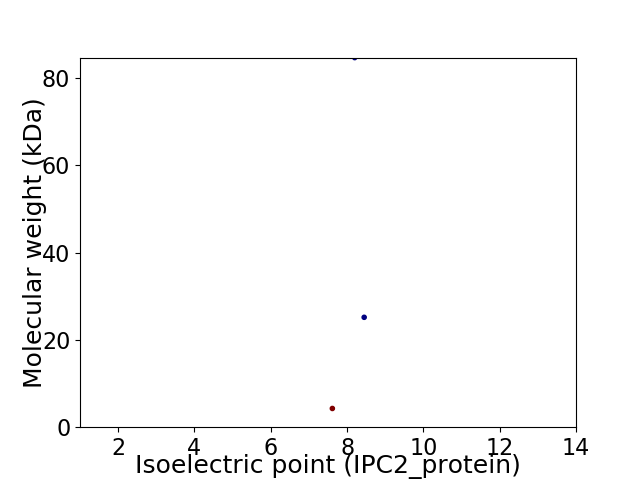
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O37365|O37365_9VIRU Uncharacterized protein OS=Carrot red leaf luteovirus associated RNA OX=67962 PE=4 SV=2
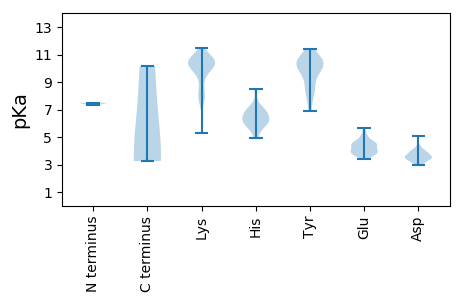
MM1 pKa = 7.49TNLNIEE7 pKa = 4.62SLVEE11 pKa = 3.87TQPAVSKK18 pKa = 10.9LVGLLPKK25 pKa = 10.47LSSRR29 pKa = 11.84LIDD32 pKa = 3.7AAKK35 pKa = 10.66VPATCGLVVAAGLGLYY51 pKa = 9.93AYY53 pKa = 10.1RR54 pKa = 11.84KK55 pKa = 8.59VCINRR60 pKa = 11.84LEE62 pKa = 4.05FSRR65 pKa = 11.84EE66 pKa = 3.66NTTPRR71 pKa = 11.84PTDD74 pKa = 3.99CIEE77 pKa = 4.52DD78 pKa = 3.6VLEE81 pKa = 4.55AEE83 pKa = 4.55EE84 pKa = 5.37DD85 pKa = 3.84AVTCLALTKK94 pKa = 9.72IAKK97 pKa = 7.77EE98 pKa = 3.9AAMEE102 pKa = 4.17AALNPGEE109 pKa = 3.71ITEE112 pKa = 4.45RR113 pKa = 11.84VRR115 pKa = 11.84EE116 pKa = 3.98AAKK119 pKa = 10.55INAQPSTDD127 pKa = 3.44PLAKK131 pKa = 9.97RR132 pKa = 11.84KK133 pKa = 7.83TRR135 pKa = 11.84LRR137 pKa = 11.84PNKK140 pKa = 8.03TGQFIRR146 pKa = 11.84VLRR149 pKa = 11.84AEE151 pKa = 4.97IKK153 pKa = 10.39SQMGTPTITAANEE166 pKa = 3.42AVIRR170 pKa = 11.84HH171 pKa = 6.13MISKK175 pKa = 9.79FCASHH180 pKa = 6.98NIRR183 pKa = 11.84TTSYY187 pKa = 8.18AHH189 pKa = 5.94LVSRR193 pKa = 11.84VVRR196 pKa = 11.84EE197 pKa = 3.73VMTPYY202 pKa = 10.43PGDD205 pKa = 3.51AEE207 pKa = 4.26EE208 pKa = 4.51VEE210 pKa = 4.53RR211 pKa = 11.84ASSVVNRR218 pKa = 11.84IHH220 pKa = 6.29NWLVQYY226 pKa = 10.53RR227 pKa = 11.84KK228 pKa = 7.98XGGLEE233 pKa = 3.66LARR236 pKa = 11.84GFTHH240 pKa = 6.94EE241 pKa = 4.57AVCTDD246 pKa = 3.52VPGLDD251 pKa = 3.41ATNLGDD257 pKa = 5.06SRR259 pKa = 11.84PLTDD263 pKa = 3.15TNIRR267 pKa = 11.84RR268 pKa = 11.84VVGPVSSDD276 pKa = 3.14YY277 pKa = 11.43DD278 pKa = 3.33IVFYY282 pKa = 10.91TNSRR286 pKa = 11.84NNLMRR291 pKa = 11.84GLVNRR296 pKa = 11.84VLTYY300 pKa = 10.52KK301 pKa = 10.51GGPVLEE307 pKa = 4.79PSPGAWKK314 pKa = 10.36SLRR317 pKa = 11.84GLATSLGNRR326 pKa = 11.84CLTTPLSPDD335 pKa = 3.27EE336 pKa = 5.13FLACYY341 pKa = 9.85VGRR344 pKa = 11.84KK345 pKa = 5.3RR346 pKa = 11.84TIYY349 pKa = 10.63AKK351 pKa = 10.38AIEE354 pKa = 4.35SFKK357 pKa = 10.18TKK359 pKa = 9.89PWDD362 pKa = 3.32ARR364 pKa = 11.84KK365 pKa = 10.8DD366 pKa = 3.69MIVKK370 pKa = 10.39AFIKK374 pKa = 10.37KK375 pKa = 10.08EE376 pKa = 3.61KK377 pKa = 10.38DD378 pKa = 3.15KK379 pKa = 11.5LVDD382 pKa = 3.74EE383 pKa = 4.92SCDD386 pKa = 3.43PRR388 pKa = 11.84IIQPRR393 pKa = 11.84TPRR396 pKa = 11.84FVTRR400 pKa = 11.84FGRR403 pKa = 11.84YY404 pKa = 6.91VRR406 pKa = 11.84AIEE409 pKa = 3.86KK410 pKa = 10.1RR411 pKa = 11.84LYY413 pKa = 10.85SEE415 pKa = 3.79WTKK418 pKa = 10.96SFSSFTVTNTPVCLKK433 pKa = 10.25GMNYY437 pKa = 7.53RR438 pKa = 11.84TRR440 pKa = 11.84ARR442 pKa = 11.84ALLEE446 pKa = 3.78KK447 pKa = 9.17WQSFDD452 pKa = 4.04SPVAVCLDD460 pKa = 3.62ASRR463 pKa = 11.84FDD465 pKa = 3.53LHH467 pKa = 8.5VSVDD471 pKa = 3.41ALKK474 pKa = 9.34FTDD477 pKa = 4.33QIYY480 pKa = 10.76LSAFTGIDD488 pKa = 3.01RR489 pKa = 11.84SEE491 pKa = 4.08LKK493 pKa = 10.36EE494 pKa = 3.77ILRR497 pKa = 11.84NRR499 pKa = 11.84HH500 pKa = 4.92KK501 pKa = 8.66TTGFATFKK509 pKa = 10.46EE510 pKa = 5.06GTFHH514 pKa = 6.14YY515 pKa = 9.25EE516 pKa = 3.81KK517 pKa = 11.08VGGRR521 pKa = 11.84CSGDD525 pKa = 3.03SDD527 pKa = 3.71TSLGNVSIMLAITRR541 pKa = 11.84VICEE545 pKa = 4.1GLKK548 pKa = 10.56GIHH551 pKa = 6.47IEE553 pKa = 4.21VANDD557 pKa = 3.44GDD559 pKa = 4.19DD560 pKa = 3.46QVLMVEE566 pKa = 4.86TNQLDD571 pKa = 3.97NLVKK575 pKa = 10.34EE576 pKa = 4.66LSPTFARR583 pKa = 11.84FGFRR587 pKa = 11.84VKK589 pKa = 10.84VEE591 pKa = 3.99DD592 pKa = 4.01PVWEE596 pKa = 4.24FEE598 pKa = 5.68RR599 pKa = 11.84IDD601 pKa = 3.62FCQTRR606 pKa = 11.84PIFLSPGEE614 pKa = 4.71PIMCRR619 pKa = 11.84YY620 pKa = 9.11PMQSMSKK627 pKa = 10.41DD628 pKa = 3.31VASFLNIEE636 pKa = 4.4RR637 pKa = 11.84SEE639 pKa = 3.9GWKK642 pKa = 10.7YY643 pKa = 8.61MLRR646 pKa = 11.84AIGACGSSSFSQVPVLGEE664 pKa = 4.1FYY666 pKa = 10.72SALSSTSNKK675 pKa = 10.49DD676 pKa = 2.94EE677 pKa = 4.45TKK679 pKa = 7.56WWRR682 pKa = 11.84RR683 pKa = 11.84TGVDD687 pKa = 3.89LGFKK691 pKa = 10.29QLTNNVGTIPYY702 pKa = 10.17DD703 pKa = 3.43EE704 pKa = 4.25VTARR708 pKa = 11.84SSFYY712 pKa = 10.37KK713 pKa = 10.73AFGILPDD720 pKa = 3.65TQAALEE726 pKa = 4.26SKK728 pKa = 9.69IRR730 pKa = 11.84NWNMSSVPHH739 pKa = 6.81IEE741 pKa = 4.01QVHH744 pKa = 5.58KK745 pKa = 10.53FSSEE749 pKa = 4.0FPVVDD754 pKa = 4.35GWAA757 pKa = 3.26
MM1 pKa = 7.49TNLNIEE7 pKa = 4.62SLVEE11 pKa = 3.87TQPAVSKK18 pKa = 10.9LVGLLPKK25 pKa = 10.47LSSRR29 pKa = 11.84LIDD32 pKa = 3.7AAKK35 pKa = 10.66VPATCGLVVAAGLGLYY51 pKa = 9.93AYY53 pKa = 10.1RR54 pKa = 11.84KK55 pKa = 8.59VCINRR60 pKa = 11.84LEE62 pKa = 4.05FSRR65 pKa = 11.84EE66 pKa = 3.66NTTPRR71 pKa = 11.84PTDD74 pKa = 3.99CIEE77 pKa = 4.52DD78 pKa = 3.6VLEE81 pKa = 4.55AEE83 pKa = 4.55EE84 pKa = 5.37DD85 pKa = 3.84AVTCLALTKK94 pKa = 9.72IAKK97 pKa = 7.77EE98 pKa = 3.9AAMEE102 pKa = 4.17AALNPGEE109 pKa = 3.71ITEE112 pKa = 4.45RR113 pKa = 11.84VRR115 pKa = 11.84EE116 pKa = 3.98AAKK119 pKa = 10.55INAQPSTDD127 pKa = 3.44PLAKK131 pKa = 9.97RR132 pKa = 11.84KK133 pKa = 7.83TRR135 pKa = 11.84LRR137 pKa = 11.84PNKK140 pKa = 8.03TGQFIRR146 pKa = 11.84VLRR149 pKa = 11.84AEE151 pKa = 4.97IKK153 pKa = 10.39SQMGTPTITAANEE166 pKa = 3.42AVIRR170 pKa = 11.84HH171 pKa = 6.13MISKK175 pKa = 9.79FCASHH180 pKa = 6.98NIRR183 pKa = 11.84TTSYY187 pKa = 8.18AHH189 pKa = 5.94LVSRR193 pKa = 11.84VVRR196 pKa = 11.84EE197 pKa = 3.73VMTPYY202 pKa = 10.43PGDD205 pKa = 3.51AEE207 pKa = 4.26EE208 pKa = 4.51VEE210 pKa = 4.53RR211 pKa = 11.84ASSVVNRR218 pKa = 11.84IHH220 pKa = 6.29NWLVQYY226 pKa = 10.53RR227 pKa = 11.84KK228 pKa = 7.98XGGLEE233 pKa = 3.66LARR236 pKa = 11.84GFTHH240 pKa = 6.94EE241 pKa = 4.57AVCTDD246 pKa = 3.52VPGLDD251 pKa = 3.41ATNLGDD257 pKa = 5.06SRR259 pKa = 11.84PLTDD263 pKa = 3.15TNIRR267 pKa = 11.84RR268 pKa = 11.84VVGPVSSDD276 pKa = 3.14YY277 pKa = 11.43DD278 pKa = 3.33IVFYY282 pKa = 10.91TNSRR286 pKa = 11.84NNLMRR291 pKa = 11.84GLVNRR296 pKa = 11.84VLTYY300 pKa = 10.52KK301 pKa = 10.51GGPVLEE307 pKa = 4.79PSPGAWKK314 pKa = 10.36SLRR317 pKa = 11.84GLATSLGNRR326 pKa = 11.84CLTTPLSPDD335 pKa = 3.27EE336 pKa = 5.13FLACYY341 pKa = 9.85VGRR344 pKa = 11.84KK345 pKa = 5.3RR346 pKa = 11.84TIYY349 pKa = 10.63AKK351 pKa = 10.38AIEE354 pKa = 4.35SFKK357 pKa = 10.18TKK359 pKa = 9.89PWDD362 pKa = 3.32ARR364 pKa = 11.84KK365 pKa = 10.8DD366 pKa = 3.69MIVKK370 pKa = 10.39AFIKK374 pKa = 10.37KK375 pKa = 10.08EE376 pKa = 3.61KK377 pKa = 10.38DD378 pKa = 3.15KK379 pKa = 11.5LVDD382 pKa = 3.74EE383 pKa = 4.92SCDD386 pKa = 3.43PRR388 pKa = 11.84IIQPRR393 pKa = 11.84TPRR396 pKa = 11.84FVTRR400 pKa = 11.84FGRR403 pKa = 11.84YY404 pKa = 6.91VRR406 pKa = 11.84AIEE409 pKa = 3.86KK410 pKa = 10.1RR411 pKa = 11.84LYY413 pKa = 10.85SEE415 pKa = 3.79WTKK418 pKa = 10.96SFSSFTVTNTPVCLKK433 pKa = 10.25GMNYY437 pKa = 7.53RR438 pKa = 11.84TRR440 pKa = 11.84ARR442 pKa = 11.84ALLEE446 pKa = 3.78KK447 pKa = 9.17WQSFDD452 pKa = 4.04SPVAVCLDD460 pKa = 3.62ASRR463 pKa = 11.84FDD465 pKa = 3.53LHH467 pKa = 8.5VSVDD471 pKa = 3.41ALKK474 pKa = 9.34FTDD477 pKa = 4.33QIYY480 pKa = 10.76LSAFTGIDD488 pKa = 3.01RR489 pKa = 11.84SEE491 pKa = 4.08LKK493 pKa = 10.36EE494 pKa = 3.77ILRR497 pKa = 11.84NRR499 pKa = 11.84HH500 pKa = 4.92KK501 pKa = 8.66TTGFATFKK509 pKa = 10.46EE510 pKa = 5.06GTFHH514 pKa = 6.14YY515 pKa = 9.25EE516 pKa = 3.81KK517 pKa = 11.08VGGRR521 pKa = 11.84CSGDD525 pKa = 3.03SDD527 pKa = 3.71TSLGNVSIMLAITRR541 pKa = 11.84VICEE545 pKa = 4.1GLKK548 pKa = 10.56GIHH551 pKa = 6.47IEE553 pKa = 4.21VANDD557 pKa = 3.44GDD559 pKa = 4.19DD560 pKa = 3.46QVLMVEE566 pKa = 4.86TNQLDD571 pKa = 3.97NLVKK575 pKa = 10.34EE576 pKa = 4.66LSPTFARR583 pKa = 11.84FGFRR587 pKa = 11.84VKK589 pKa = 10.84VEE591 pKa = 3.99DD592 pKa = 4.01PVWEE596 pKa = 4.24FEE598 pKa = 5.68RR599 pKa = 11.84IDD601 pKa = 3.62FCQTRR606 pKa = 11.84PIFLSPGEE614 pKa = 4.71PIMCRR619 pKa = 11.84YY620 pKa = 9.11PMQSMSKK627 pKa = 10.41DD628 pKa = 3.31VASFLNIEE636 pKa = 4.4RR637 pKa = 11.84SEE639 pKa = 3.9GWKK642 pKa = 10.7YY643 pKa = 8.61MLRR646 pKa = 11.84AIGACGSSSFSQVPVLGEE664 pKa = 4.1FYY666 pKa = 10.72SALSSTSNKK675 pKa = 10.49DD676 pKa = 2.94EE677 pKa = 4.45TKK679 pKa = 7.56WWRR682 pKa = 11.84RR683 pKa = 11.84TGVDD687 pKa = 3.89LGFKK691 pKa = 10.29QLTNNVGTIPYY702 pKa = 10.17DD703 pKa = 3.43EE704 pKa = 4.25VTARR708 pKa = 11.84SSFYY712 pKa = 10.37KK713 pKa = 10.73AFGILPDD720 pKa = 3.65TQAALEE726 pKa = 4.26SKK728 pKa = 9.69IRR730 pKa = 11.84NWNMSSVPHH739 pKa = 6.81IEE741 pKa = 4.01QVHH744 pKa = 5.58KK745 pKa = 10.53FSSEE749 pKa = 4.0FPVVDD754 pKa = 4.35GWAA757 pKa = 3.26
Molecular weight: 84.68 kDa
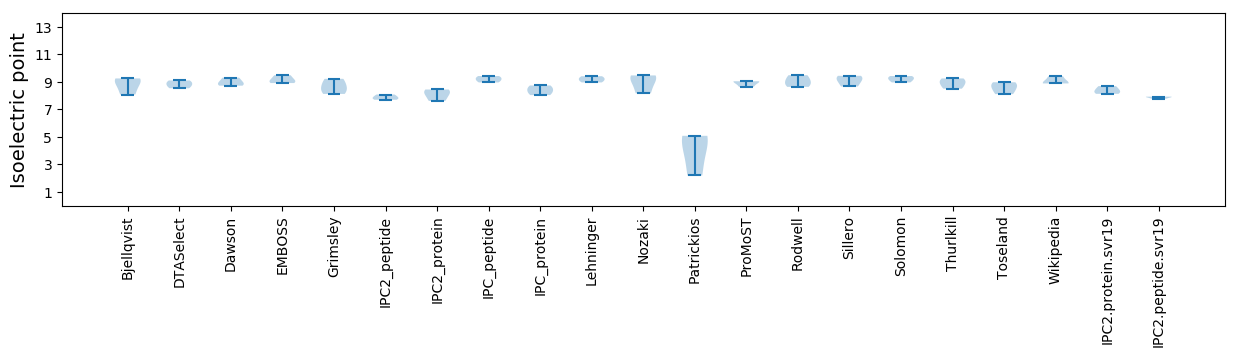
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O37366|O37366_9VIRU Uncharacterized protein OS=Carrot red leaf luteovirus associated RNA OX=67962 PE=4 SV=1
MM1 pKa = 7.49TNLNIEE7 pKa = 4.62SLVEE11 pKa = 3.87TQPAVSKK18 pKa = 10.9LVGLLPKK25 pKa = 10.47LSSRR29 pKa = 11.84LIDD32 pKa = 3.7AAKK35 pKa = 10.66VPATCGLVVAAGLGLYY51 pKa = 9.93AYY53 pKa = 10.1RR54 pKa = 11.84KK55 pKa = 8.59VCINRR60 pKa = 11.84LEE62 pKa = 4.05FSRR65 pKa = 11.84EE66 pKa = 3.66NTTPRR71 pKa = 11.84PTDD74 pKa = 3.99CIEE77 pKa = 4.52DD78 pKa = 3.6VLEE81 pKa = 4.55AEE83 pKa = 4.55EE84 pKa = 5.37DD85 pKa = 3.84AVTCLALTKK94 pKa = 9.72IAKK97 pKa = 7.77EE98 pKa = 3.9AAMEE102 pKa = 4.17AALNPGEE109 pKa = 3.71ITEE112 pKa = 4.45RR113 pKa = 11.84VRR115 pKa = 11.84EE116 pKa = 3.98AAKK119 pKa = 10.55INAQPSTDD127 pKa = 3.44PLAKK131 pKa = 9.97RR132 pKa = 11.84KK133 pKa = 7.83TRR135 pKa = 11.84LRR137 pKa = 11.84PNKK140 pKa = 8.03TGQFIRR146 pKa = 11.84VLRR149 pKa = 11.84AEE151 pKa = 4.97IKK153 pKa = 10.39SQMGTPTITAANEE166 pKa = 3.42AVIRR170 pKa = 11.84HH171 pKa = 6.13MISKK175 pKa = 9.79FCASHH180 pKa = 6.98NIRR183 pKa = 11.84TTSYY187 pKa = 8.18AHH189 pKa = 5.94LVSRR193 pKa = 11.84VVRR196 pKa = 11.84EE197 pKa = 3.73VMTPYY202 pKa = 10.43PGDD205 pKa = 3.51AEE207 pKa = 4.26EE208 pKa = 4.51VEE210 pKa = 4.53RR211 pKa = 11.84ASSVVNRR218 pKa = 11.84IHH220 pKa = 6.29NWLVQYY226 pKa = 10.79RR227 pKa = 11.84KK228 pKa = 10.18
MM1 pKa = 7.49TNLNIEE7 pKa = 4.62SLVEE11 pKa = 3.87TQPAVSKK18 pKa = 10.9LVGLLPKK25 pKa = 10.47LSSRR29 pKa = 11.84LIDD32 pKa = 3.7AAKK35 pKa = 10.66VPATCGLVVAAGLGLYY51 pKa = 9.93AYY53 pKa = 10.1RR54 pKa = 11.84KK55 pKa = 8.59VCINRR60 pKa = 11.84LEE62 pKa = 4.05FSRR65 pKa = 11.84EE66 pKa = 3.66NTTPRR71 pKa = 11.84PTDD74 pKa = 3.99CIEE77 pKa = 4.52DD78 pKa = 3.6VLEE81 pKa = 4.55AEE83 pKa = 4.55EE84 pKa = 5.37DD85 pKa = 3.84AVTCLALTKK94 pKa = 9.72IAKK97 pKa = 7.77EE98 pKa = 3.9AAMEE102 pKa = 4.17AALNPGEE109 pKa = 3.71ITEE112 pKa = 4.45RR113 pKa = 11.84VRR115 pKa = 11.84EE116 pKa = 3.98AAKK119 pKa = 10.55INAQPSTDD127 pKa = 3.44PLAKK131 pKa = 9.97RR132 pKa = 11.84KK133 pKa = 7.83TRR135 pKa = 11.84LRR137 pKa = 11.84PNKK140 pKa = 8.03TGQFIRR146 pKa = 11.84VLRR149 pKa = 11.84AEE151 pKa = 4.97IKK153 pKa = 10.39SQMGTPTITAANEE166 pKa = 3.42AVIRR170 pKa = 11.84HH171 pKa = 6.13MISKK175 pKa = 9.79FCASHH180 pKa = 6.98NIRR183 pKa = 11.84TTSYY187 pKa = 8.18AHH189 pKa = 5.94LVSRR193 pKa = 11.84VVRR196 pKa = 11.84EE197 pKa = 3.73VMTPYY202 pKa = 10.43PGDD205 pKa = 3.51AEE207 pKa = 4.26EE208 pKa = 4.51VEE210 pKa = 4.53RR211 pKa = 11.84ASSVVNRR218 pKa = 11.84IHH220 pKa = 6.29NWLVQYY226 pKa = 10.79RR227 pKa = 11.84KK228 pKa = 10.18
Molecular weight: 25.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1024 |
39 |
757 |
341.3 |
38.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.301 ± 1.861 | 2.051 ± 0.461 |
4.492 ± 0.806 | 6.738 ± 1.085 |
3.516 ± 1.351 | 5.762 ± 2.789 |
1.563 ± 0.256 | 5.371 ± 0.672 |
5.859 ± 0.19 | 8.203 ± 0.704 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.051 ± 0.141 | 4.199 ± 0.425 |
5.176 ± 1.163 | 2.051 ± 0.461 |
7.715 ± 0.607 | 7.227 ± 0.66 |
7.324 ± 0.521 | 8.105 ± 0.292 |
1.27 ± 0.43 | 2.93 ± 2.222 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
