
Tessaracoccus rhinocerotis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Tessaracoccus
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
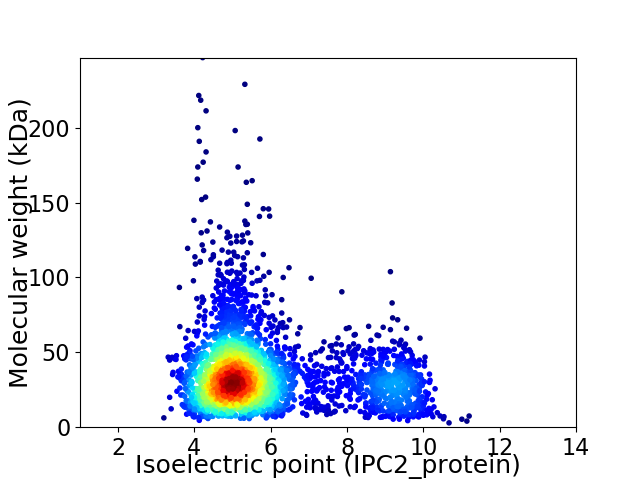
Virtual 2D-PAGE plot for 3082 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A553JZN4|A0A553JZN4_9ACTN ATP-binding protein OS=Tessaracoccus rhinocerotis OX=1689449 GN=FOJ82_11680 PE=4 SV=1
MM1 pKa = 7.24NRR3 pKa = 11.84RR4 pKa = 11.84LIAASAALASLALLVTGCGADD25 pKa = 3.58TEE27 pKa = 4.98APTDD31 pKa = 3.68TEE33 pKa = 4.44TTAGGTTAAEE43 pKa = 4.14TTPAGDD49 pKa = 3.52TGSTEE54 pKa = 4.26PKK56 pKa = 10.83GEE58 pKa = 3.97LTLAGWSLNTTPEE71 pKa = 4.04FQTLADD77 pKa = 4.46GFNATNPEE85 pKa = 4.04YY86 pKa = 9.93TVTVSEE92 pKa = 4.32YY93 pKa = 10.49QAGNDD98 pKa = 3.9YY99 pKa = 8.41DD100 pKa = 3.94TQMITDD106 pKa = 4.92LAAGTAPDD114 pKa = 4.46LYY116 pKa = 10.75IMKK119 pKa = 10.05NLVNFYY125 pKa = 9.75TYY127 pKa = 11.09AAGEE131 pKa = 4.08QLVDD135 pKa = 3.66VSDD138 pKa = 3.97VAGTLDD144 pKa = 3.69NEE146 pKa = 4.64SVSAYY151 pKa = 9.5EE152 pKa = 4.86LDD154 pKa = 3.48GATYY158 pKa = 9.71AIPYY162 pKa = 9.3RR163 pKa = 11.84QDD165 pKa = 2.41AWFLYY170 pKa = 10.07YY171 pKa = 10.83NIDD174 pKa = 3.56LFEE177 pKa = 4.1QAGVEE182 pKa = 4.32VPDD185 pKa = 6.1GSWTWDD191 pKa = 3.5DD192 pKa = 3.49YY193 pKa = 11.95AAAATEE199 pKa = 4.05LSEE202 pKa = 4.97KK203 pKa = 11.13LEE205 pKa = 4.33GDD207 pKa = 3.4TKK209 pKa = 10.97GAYY212 pKa = 4.97THH214 pKa = 6.43SWQSVIQGFANAQSEE229 pKa = 4.55GADD232 pKa = 3.67VLSGEE237 pKa = 4.25YY238 pKa = 10.45DD239 pKa = 3.24HH240 pKa = 7.04LKK242 pKa = 10.31PYY244 pKa = 9.64YY245 pKa = 10.01EE246 pKa = 5.21RR247 pKa = 11.84ALQMQADD254 pKa = 4.52GAMEE258 pKa = 4.72AYY260 pKa = 8.59GTVTTNSLSYY270 pKa = 10.33QSQFGTQKK278 pKa = 11.26AAMLPMGSWYY288 pKa = 9.95IATLVAQQASGEE300 pKa = 4.26AEE302 pKa = 3.89DD303 pKa = 4.59FAWGIAPAPQFDD315 pKa = 4.39EE316 pKa = 4.69STAGTDD322 pKa = 3.33NTPVTFGDD330 pKa = 3.58PTGIGINPAIEE341 pKa = 4.28DD342 pKa = 4.07EE343 pKa = 4.54KK344 pKa = 11.48LDD346 pKa = 3.64TAKK349 pKa = 11.1AFLSYY354 pKa = 10.44VASEE358 pKa = 4.39EE359 pKa = 4.28GAKK362 pKa = 10.39ALAGIGITPSVMSPEE377 pKa = 4.05VTDD380 pKa = 6.15LFFQLDD386 pKa = 4.86GIAEE390 pKa = 4.49DD391 pKa = 4.28EE392 pKa = 4.21LSRR395 pKa = 11.84WTFEE399 pKa = 3.76NRR401 pKa = 11.84TVMPEE406 pKa = 3.75NSVSQHH412 pKa = 4.06TAEE415 pKa = 4.35INTLLGEE422 pKa = 4.07LHH424 pKa = 6.5SAVLSDD430 pKa = 5.1SEE432 pKa = 5.05GIDD435 pKa = 3.32AAIEE439 pKa = 4.01NAEE442 pKa = 3.84NRR444 pKa = 11.84AKK446 pKa = 11.04NEE448 pKa = 3.56ILNRR452 pKa = 3.9
MM1 pKa = 7.24NRR3 pKa = 11.84RR4 pKa = 11.84LIAASAALASLALLVTGCGADD25 pKa = 3.58TEE27 pKa = 4.98APTDD31 pKa = 3.68TEE33 pKa = 4.44TTAGGTTAAEE43 pKa = 4.14TTPAGDD49 pKa = 3.52TGSTEE54 pKa = 4.26PKK56 pKa = 10.83GEE58 pKa = 3.97LTLAGWSLNTTPEE71 pKa = 4.04FQTLADD77 pKa = 4.46GFNATNPEE85 pKa = 4.04YY86 pKa = 9.93TVTVSEE92 pKa = 4.32YY93 pKa = 10.49QAGNDD98 pKa = 3.9YY99 pKa = 8.41DD100 pKa = 3.94TQMITDD106 pKa = 4.92LAAGTAPDD114 pKa = 4.46LYY116 pKa = 10.75IMKK119 pKa = 10.05NLVNFYY125 pKa = 9.75TYY127 pKa = 11.09AAGEE131 pKa = 4.08QLVDD135 pKa = 3.66VSDD138 pKa = 3.97VAGTLDD144 pKa = 3.69NEE146 pKa = 4.64SVSAYY151 pKa = 9.5EE152 pKa = 4.86LDD154 pKa = 3.48GATYY158 pKa = 9.71AIPYY162 pKa = 9.3RR163 pKa = 11.84QDD165 pKa = 2.41AWFLYY170 pKa = 10.07YY171 pKa = 10.83NIDD174 pKa = 3.56LFEE177 pKa = 4.1QAGVEE182 pKa = 4.32VPDD185 pKa = 6.1GSWTWDD191 pKa = 3.5DD192 pKa = 3.49YY193 pKa = 11.95AAAATEE199 pKa = 4.05LSEE202 pKa = 4.97KK203 pKa = 11.13LEE205 pKa = 4.33GDD207 pKa = 3.4TKK209 pKa = 10.97GAYY212 pKa = 4.97THH214 pKa = 6.43SWQSVIQGFANAQSEE229 pKa = 4.55GADD232 pKa = 3.67VLSGEE237 pKa = 4.25YY238 pKa = 10.45DD239 pKa = 3.24HH240 pKa = 7.04LKK242 pKa = 10.31PYY244 pKa = 9.64YY245 pKa = 10.01EE246 pKa = 5.21RR247 pKa = 11.84ALQMQADD254 pKa = 4.52GAMEE258 pKa = 4.72AYY260 pKa = 8.59GTVTTNSLSYY270 pKa = 10.33QSQFGTQKK278 pKa = 11.26AAMLPMGSWYY288 pKa = 9.95IATLVAQQASGEE300 pKa = 4.26AEE302 pKa = 3.89DD303 pKa = 4.59FAWGIAPAPQFDD315 pKa = 4.39EE316 pKa = 4.69STAGTDD322 pKa = 3.33NTPVTFGDD330 pKa = 3.58PTGIGINPAIEE341 pKa = 4.28DD342 pKa = 4.07EE343 pKa = 4.54KK344 pKa = 11.48LDD346 pKa = 3.64TAKK349 pKa = 11.1AFLSYY354 pKa = 10.44VASEE358 pKa = 4.39EE359 pKa = 4.28GAKK362 pKa = 10.39ALAGIGITPSVMSPEE377 pKa = 4.05VTDD380 pKa = 6.15LFFQLDD386 pKa = 4.86GIAEE390 pKa = 4.49DD391 pKa = 4.28EE392 pKa = 4.21LSRR395 pKa = 11.84WTFEE399 pKa = 3.76NRR401 pKa = 11.84TVMPEE406 pKa = 3.75NSVSQHH412 pKa = 4.06TAEE415 pKa = 4.35INTLLGEE422 pKa = 4.07LHH424 pKa = 6.5SAVLSDD430 pKa = 5.1SEE432 pKa = 5.05GIDD435 pKa = 3.32AAIEE439 pKa = 4.01NAEE442 pKa = 3.84NRR444 pKa = 11.84AKK446 pKa = 11.04NEE448 pKa = 3.56ILNRR452 pKa = 3.9
Molecular weight: 48.29 kDa
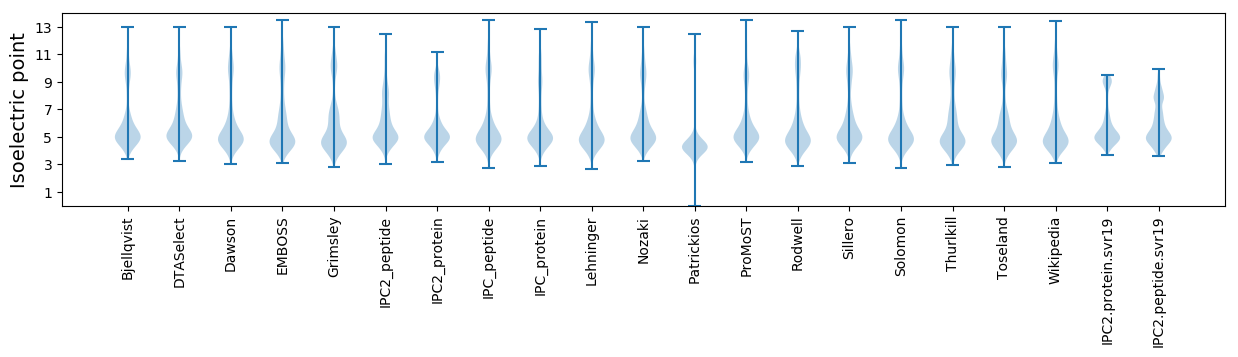
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A553JX90|A0A553JX90_9ACTN Nucleoside/nucleotide kinase family protein OS=Tessaracoccus rhinocerotis OX=1689449 GN=FOJ82_14640 PE=4 SV=1
MM1 pKa = 7.49VFQQGAAPARR11 pKa = 11.84GTPGQPFRR19 pKa = 11.84GSDD22 pKa = 2.95AGRR25 pKa = 11.84VSRR28 pKa = 11.84HH29 pKa = 3.98RR30 pKa = 11.84RR31 pKa = 11.84HH32 pKa = 6.26RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SHH37 pKa = 5.31HH38 pKa = 5.62RR39 pKa = 11.84RR40 pKa = 11.84HH41 pKa = 5.99RR42 pKa = 11.84VRR44 pKa = 11.84PSRR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 4.34RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84PARR57 pKa = 11.84AAAVGPAA64 pKa = 3.41
MM1 pKa = 7.49VFQQGAAPARR11 pKa = 11.84GTPGQPFRR19 pKa = 11.84GSDD22 pKa = 2.95AGRR25 pKa = 11.84VSRR28 pKa = 11.84HH29 pKa = 3.98RR30 pKa = 11.84RR31 pKa = 11.84HH32 pKa = 6.26RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SHH37 pKa = 5.31HH38 pKa = 5.62RR39 pKa = 11.84RR40 pKa = 11.84HH41 pKa = 5.99RR42 pKa = 11.84VRR44 pKa = 11.84PSRR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 4.34RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84PARR57 pKa = 11.84AAAVGPAA64 pKa = 3.41
Molecular weight: 7.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1069584 |
24 |
2321 |
347.0 |
37.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.196 ± 0.056 | 0.661 ± 0.01 |
6.151 ± 0.035 | 6.207 ± 0.043 |
3.165 ± 0.027 | 9.056 ± 0.036 |
2.19 ± 0.021 | 3.978 ± 0.035 |
2.074 ± 0.028 | 10.119 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.018 ± 0.02 | 2.3 ± 0.028 |
5.439 ± 0.033 | 3.01 ± 0.022 |
7.128 ± 0.05 | 5.535 ± 0.029 |
6.017 ± 0.037 | 9.091 ± 0.039 |
1.671 ± 0.023 | 1.993 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
