
Enhydrobacter sp. CC-CFT640
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillales incertae sedis; Enhydrobacter; unclassified Enhydrobacter
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
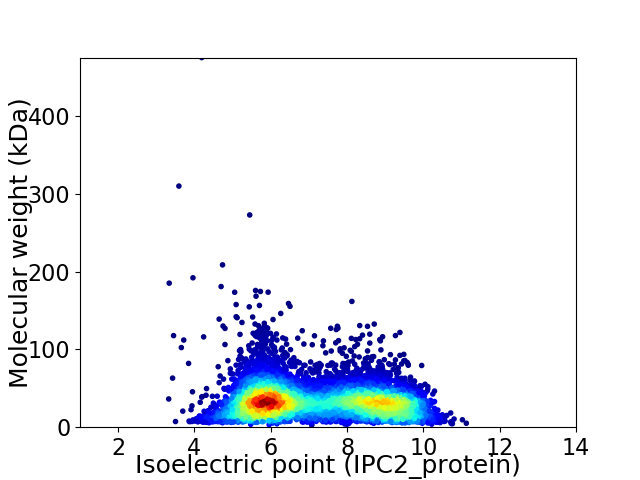
Virtual 2D-PAGE plot for 7888 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C8PBN7|A0A5C8PBN7_9PROT Uncharacterized protein OS=Enhydrobacter sp. CC-CFT640 OX=2586908 GN=FHP25_32560 PE=4 SV=1
MM1 pKa = 7.56AGDD4 pKa = 4.68GSTGAPGGTPQVPHH18 pKa = 7.24LLDD21 pKa = 4.74GYY23 pKa = 10.23AVRR26 pKa = 11.84PAWEE30 pKa = 4.13VAGVDD35 pKa = 3.87YY36 pKa = 11.08AVGVHH41 pKa = 6.3PGITLRR47 pKa = 11.84VPTADD52 pKa = 3.75NLPAGTTLGNHH63 pKa = 6.55VIYY66 pKa = 10.63INGTDD71 pKa = 3.61VTLSGYY77 pKa = 10.7NLTNYY82 pKa = 7.95TVLVTATAKK91 pKa = 9.21GTATIVDD98 pKa = 4.39CAATTGINIRR108 pKa = 11.84STVDD112 pKa = 2.8ATANLVVRR120 pKa = 11.84YY121 pKa = 9.65CSFDD125 pKa = 3.39GGGMASDD132 pKa = 4.81PDD134 pKa = 3.89FSLIKK139 pKa = 9.66VWCPLTVEE147 pKa = 4.01YY148 pKa = 10.73SVLKK152 pKa = 10.07NAPAAIYY159 pKa = 9.99APEE162 pKa = 4.73PMTVMYY168 pKa = 11.09NVMSGFAWQDD178 pKa = 3.06GAHH181 pKa = 6.46ACAIAVEE188 pKa = 4.68GTNDD192 pKa = 3.5PNASALIAYY201 pKa = 7.18NTIYY205 pKa = 10.93SGDD208 pKa = 3.37ARR210 pKa = 11.84NAEE213 pKa = 4.28GFPIGIGTGIAFYY226 pKa = 11.17NDD228 pKa = 3.0AGGNFYY234 pKa = 11.17NSTVANNTVIANLPGGASYY253 pKa = 9.08LTGFYY258 pKa = 10.57VDD260 pKa = 4.25PPGTATNMHH269 pKa = 5.74VQDD272 pKa = 3.76NFYY275 pKa = 11.34ASVNGFNNASSGAYY289 pKa = 7.87GALYY293 pKa = 10.02IGTKK297 pKa = 9.24GTVQATYY304 pKa = 10.2TNNIDD309 pKa = 3.37MSTGQLVAGSDD320 pKa = 3.75AGPVDD325 pKa = 3.67TGTVQSDD332 pKa = 3.22VSYY335 pKa = 9.15TLPAGIEE342 pKa = 4.05NLTLTGSDD350 pKa = 4.02NINGTGNSLANVLTGNAGNNILDD373 pKa = 3.92GGANADD379 pKa = 3.91HH380 pKa = 6.61MAGGAGDD387 pKa = 3.72DD388 pKa = 3.98TYY390 pKa = 11.99VVDD393 pKa = 3.67NAGDD397 pKa = 3.74VVVEE401 pKa = 4.27EE402 pKa = 4.63NGQGTDD408 pKa = 3.32TVLSSVSYY416 pKa = 11.05SLANYY421 pKa = 9.66IEE423 pKa = 4.29NLTLTGNGNINGTGNSLANVLTGNSGSNTLNGGLGADD460 pKa = 3.89TLNGGGGNDD469 pKa = 3.93TLNGGAGNDD478 pKa = 3.99TLNGGAGNDD487 pKa = 3.56TAVYY491 pKa = 9.9AGNRR495 pKa = 11.84ADD497 pKa = 4.07YY498 pKa = 10.35EE499 pKa = 4.08ILAVGGGLTIRR510 pKa = 11.84DD511 pKa = 4.06LNPANGDD518 pKa = 3.55EE519 pKa = 4.68GTDD522 pKa = 3.41TLQGIEE528 pKa = 4.22MLRR531 pKa = 11.84FADD534 pKa = 3.29ITVPASTSNAAPVAGDD550 pKa = 3.84DD551 pKa = 3.94SASGSEE557 pKa = 4.42DD558 pKa = 3.21KK559 pKa = 11.1AITTGNVLANDD570 pKa = 3.69SDD572 pKa = 4.32ADD574 pKa = 3.93STLTAASIIGFSQGAHH590 pKa = 5.52GAVVYY595 pKa = 10.61NGNGTFTYY603 pKa = 10.29TPVPNYY609 pKa = 10.57NGADD613 pKa = 3.25SFTYY617 pKa = 9.56TVSDD621 pKa = 3.31GHH623 pKa = 6.5GGSDD627 pKa = 3.42TARR630 pKa = 11.84VSITVNPVNDD640 pKa = 3.9APVAIDD646 pKa = 5.18DD647 pKa = 5.07GPLQATGDD655 pKa = 3.73QPLVISAAALLANDD669 pKa = 3.99TDD671 pKa = 4.29ADD673 pKa = 4.27LDD675 pKa = 3.77ALTIQGVTQPAHH687 pKa = 6.41GALADD692 pKa = 3.83NGNGTFTYY700 pKa = 10.27TPVRR704 pKa = 11.84NYY706 pKa = 10.89NGADD710 pKa = 3.02SFTYY714 pKa = 9.56TVSDD718 pKa = 3.31GHH720 pKa = 6.5GGSDD724 pKa = 3.42TARR727 pKa = 11.84VSIAVNPVNDD737 pKa = 4.07APVAIDD743 pKa = 5.18DD744 pKa = 5.07GPLQATGDD752 pKa = 3.73QPLVISAAALLANDD766 pKa = 3.82TDD768 pKa = 4.29ADD770 pKa = 3.92FDD772 pKa = 4.15ALTIQGVTQPTHH784 pKa = 5.42GTLADD789 pKa = 3.72NGNGTFTYY797 pKa = 10.0TRR799 pKa = 11.84DD800 pKa = 3.35AGYY803 pKa = 10.92VGADD807 pKa = 2.98SFTYY811 pKa = 10.13TVDD814 pKa = 3.88DD815 pKa = 3.85GHH817 pKa = 6.8GGSDD821 pKa = 3.2TATVSIRR828 pKa = 11.84VDD830 pKa = 3.34DD831 pKa = 4.07AAGRR835 pKa = 11.84TLTGNEE841 pKa = 3.69RR842 pKa = 11.84ANYY845 pKa = 9.76LVGGAGNDD853 pKa = 3.71TLNGRR858 pKa = 11.84GGNDD862 pKa = 3.27TLNGRR867 pKa = 11.84GGNDD871 pKa = 3.28TLNGGGGNDD880 pKa = 3.81TLNGGDD886 pKa = 5.4GDD888 pKa = 4.04DD889 pKa = 4.33TLNGGAGNDD898 pKa = 3.59TLLGGRR904 pKa = 11.84GRR906 pKa = 11.84DD907 pKa = 3.64VLIGGAGADD916 pKa = 3.81LFDD919 pKa = 3.66FHH921 pKa = 8.18AFNEE925 pKa = 4.52SGTTDD930 pKa = 3.06ATRR933 pKa = 11.84DD934 pKa = 3.49QIVGFEE940 pKa = 4.14RR941 pKa = 11.84GTDD944 pKa = 3.56RR945 pKa = 11.84IDD947 pKa = 4.27LATIDD952 pKa = 4.63ANTAAAGNQAFAFIGGAAFHH972 pKa = 6.39GVAGEE977 pKa = 4.12LRR979 pKa = 11.84QQAFGADD986 pKa = 3.83TIVSGDD992 pKa = 3.58VNGDD996 pKa = 3.25AVADD1000 pKa = 3.94FQIQLKK1006 pKa = 10.4GAFALTASDD1015 pKa = 4.73FFLL1018 pKa = 5.94
MM1 pKa = 7.56AGDD4 pKa = 4.68GSTGAPGGTPQVPHH18 pKa = 7.24LLDD21 pKa = 4.74GYY23 pKa = 10.23AVRR26 pKa = 11.84PAWEE30 pKa = 4.13VAGVDD35 pKa = 3.87YY36 pKa = 11.08AVGVHH41 pKa = 6.3PGITLRR47 pKa = 11.84VPTADD52 pKa = 3.75NLPAGTTLGNHH63 pKa = 6.55VIYY66 pKa = 10.63INGTDD71 pKa = 3.61VTLSGYY77 pKa = 10.7NLTNYY82 pKa = 7.95TVLVTATAKK91 pKa = 9.21GTATIVDD98 pKa = 4.39CAATTGINIRR108 pKa = 11.84STVDD112 pKa = 2.8ATANLVVRR120 pKa = 11.84YY121 pKa = 9.65CSFDD125 pKa = 3.39GGGMASDD132 pKa = 4.81PDD134 pKa = 3.89FSLIKK139 pKa = 9.66VWCPLTVEE147 pKa = 4.01YY148 pKa = 10.73SVLKK152 pKa = 10.07NAPAAIYY159 pKa = 9.99APEE162 pKa = 4.73PMTVMYY168 pKa = 11.09NVMSGFAWQDD178 pKa = 3.06GAHH181 pKa = 6.46ACAIAVEE188 pKa = 4.68GTNDD192 pKa = 3.5PNASALIAYY201 pKa = 7.18NTIYY205 pKa = 10.93SGDD208 pKa = 3.37ARR210 pKa = 11.84NAEE213 pKa = 4.28GFPIGIGTGIAFYY226 pKa = 11.17NDD228 pKa = 3.0AGGNFYY234 pKa = 11.17NSTVANNTVIANLPGGASYY253 pKa = 9.08LTGFYY258 pKa = 10.57VDD260 pKa = 4.25PPGTATNMHH269 pKa = 5.74VQDD272 pKa = 3.76NFYY275 pKa = 11.34ASVNGFNNASSGAYY289 pKa = 7.87GALYY293 pKa = 10.02IGTKK297 pKa = 9.24GTVQATYY304 pKa = 10.2TNNIDD309 pKa = 3.37MSTGQLVAGSDD320 pKa = 3.75AGPVDD325 pKa = 3.67TGTVQSDD332 pKa = 3.22VSYY335 pKa = 9.15TLPAGIEE342 pKa = 4.05NLTLTGSDD350 pKa = 4.02NINGTGNSLANVLTGNAGNNILDD373 pKa = 3.92GGANADD379 pKa = 3.91HH380 pKa = 6.61MAGGAGDD387 pKa = 3.72DD388 pKa = 3.98TYY390 pKa = 11.99VVDD393 pKa = 3.67NAGDD397 pKa = 3.74VVVEE401 pKa = 4.27EE402 pKa = 4.63NGQGTDD408 pKa = 3.32TVLSSVSYY416 pKa = 11.05SLANYY421 pKa = 9.66IEE423 pKa = 4.29NLTLTGNGNINGTGNSLANVLTGNSGSNTLNGGLGADD460 pKa = 3.89TLNGGGGNDD469 pKa = 3.93TLNGGAGNDD478 pKa = 3.99TLNGGAGNDD487 pKa = 3.56TAVYY491 pKa = 9.9AGNRR495 pKa = 11.84ADD497 pKa = 4.07YY498 pKa = 10.35EE499 pKa = 4.08ILAVGGGLTIRR510 pKa = 11.84DD511 pKa = 4.06LNPANGDD518 pKa = 3.55EE519 pKa = 4.68GTDD522 pKa = 3.41TLQGIEE528 pKa = 4.22MLRR531 pKa = 11.84FADD534 pKa = 3.29ITVPASTSNAAPVAGDD550 pKa = 3.84DD551 pKa = 3.94SASGSEE557 pKa = 4.42DD558 pKa = 3.21KK559 pKa = 11.1AITTGNVLANDD570 pKa = 3.69SDD572 pKa = 4.32ADD574 pKa = 3.93STLTAASIIGFSQGAHH590 pKa = 5.52GAVVYY595 pKa = 10.61NGNGTFTYY603 pKa = 10.29TPVPNYY609 pKa = 10.57NGADD613 pKa = 3.25SFTYY617 pKa = 9.56TVSDD621 pKa = 3.31GHH623 pKa = 6.5GGSDD627 pKa = 3.42TARR630 pKa = 11.84VSITVNPVNDD640 pKa = 3.9APVAIDD646 pKa = 5.18DD647 pKa = 5.07GPLQATGDD655 pKa = 3.73QPLVISAAALLANDD669 pKa = 3.99TDD671 pKa = 4.29ADD673 pKa = 4.27LDD675 pKa = 3.77ALTIQGVTQPAHH687 pKa = 6.41GALADD692 pKa = 3.83NGNGTFTYY700 pKa = 10.27TPVRR704 pKa = 11.84NYY706 pKa = 10.89NGADD710 pKa = 3.02SFTYY714 pKa = 9.56TVSDD718 pKa = 3.31GHH720 pKa = 6.5GGSDD724 pKa = 3.42TARR727 pKa = 11.84VSIAVNPVNDD737 pKa = 4.07APVAIDD743 pKa = 5.18DD744 pKa = 5.07GPLQATGDD752 pKa = 3.73QPLVISAAALLANDD766 pKa = 3.82TDD768 pKa = 4.29ADD770 pKa = 3.92FDD772 pKa = 4.15ALTIQGVTQPTHH784 pKa = 5.42GTLADD789 pKa = 3.72NGNGTFTYY797 pKa = 10.0TRR799 pKa = 11.84DD800 pKa = 3.35AGYY803 pKa = 10.92VGADD807 pKa = 2.98SFTYY811 pKa = 10.13TVDD814 pKa = 3.88DD815 pKa = 3.85GHH817 pKa = 6.8GGSDD821 pKa = 3.2TATVSIRR828 pKa = 11.84VDD830 pKa = 3.34DD831 pKa = 4.07AAGRR835 pKa = 11.84TLTGNEE841 pKa = 3.69RR842 pKa = 11.84ANYY845 pKa = 9.76LVGGAGNDD853 pKa = 3.71TLNGRR858 pKa = 11.84GGNDD862 pKa = 3.27TLNGRR867 pKa = 11.84GGNDD871 pKa = 3.28TLNGGGGNDD880 pKa = 3.81TLNGGDD886 pKa = 5.4GDD888 pKa = 4.04DD889 pKa = 4.33TLNGGAGNDD898 pKa = 3.59TLLGGRR904 pKa = 11.84GRR906 pKa = 11.84DD907 pKa = 3.64VLIGGAGADD916 pKa = 3.81LFDD919 pKa = 3.66FHH921 pKa = 8.18AFNEE925 pKa = 4.52SGTTDD930 pKa = 3.06ATRR933 pKa = 11.84DD934 pKa = 3.49QIVGFEE940 pKa = 4.14RR941 pKa = 11.84GTDD944 pKa = 3.56RR945 pKa = 11.84IDD947 pKa = 4.27LATIDD952 pKa = 4.63ANTAAAGNQAFAFIGGAAFHH972 pKa = 6.39GVAGEE977 pKa = 4.12LRR979 pKa = 11.84QQAFGADD986 pKa = 3.83TIVSGDD992 pKa = 3.58VNGDD996 pKa = 3.25AVADD1000 pKa = 3.94FQIQLKK1006 pKa = 10.4GAFALTASDD1015 pKa = 4.73FFLL1018 pKa = 5.94
Molecular weight: 102.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C8P6U6|A0A5C8P6U6_9PROT Adenylate/guanylate cyclase domain-containing protein OS=Enhydrobacter sp. CC-CFT640 OX=2586908 GN=FHP25_39895 PE=4 SV=1
MM1 pKa = 7.87AKK3 pKa = 8.84RR4 pKa = 11.84TTKK7 pKa = 10.6SAVARR12 pKa = 11.84TKK14 pKa = 8.25TAKK17 pKa = 10.37KK18 pKa = 9.33KK19 pKa = 9.62VEE21 pKa = 4.07RR22 pKa = 11.84TTVTTKK28 pKa = 10.42RR29 pKa = 11.84RR30 pKa = 11.84TAAKK34 pKa = 9.82AKK36 pKa = 10.0VKK38 pKa = 10.77AKK40 pKa = 10.78VKK42 pKa = 10.64VKK44 pKa = 10.33VKK46 pKa = 10.28KK47 pKa = 9.25AAKK50 pKa = 9.57RR51 pKa = 11.84VAPKK55 pKa = 10.24AAAKK59 pKa = 9.83SARR62 pKa = 11.84AGTAVKK68 pKa = 10.37RR69 pKa = 11.84PAAKK73 pKa = 9.98KK74 pKa = 9.59RR75 pKa = 11.84VAAKK79 pKa = 10.12RR80 pKa = 11.84ATSKK84 pKa = 9.36TRR86 pKa = 11.84ASRR89 pKa = 3.35
MM1 pKa = 7.87AKK3 pKa = 8.84RR4 pKa = 11.84TTKK7 pKa = 10.6SAVARR12 pKa = 11.84TKK14 pKa = 8.25TAKK17 pKa = 10.37KK18 pKa = 9.33KK19 pKa = 9.62VEE21 pKa = 4.07RR22 pKa = 11.84TTVTTKK28 pKa = 10.42RR29 pKa = 11.84RR30 pKa = 11.84TAAKK34 pKa = 9.82AKK36 pKa = 10.0VKK38 pKa = 10.77AKK40 pKa = 10.78VKK42 pKa = 10.64VKK44 pKa = 10.33VKK46 pKa = 10.28KK47 pKa = 9.25AAKK50 pKa = 9.57RR51 pKa = 11.84VAPKK55 pKa = 10.24AAAKK59 pKa = 9.83SARR62 pKa = 11.84AGTAVKK68 pKa = 10.37RR69 pKa = 11.84PAAKK73 pKa = 9.98KK74 pKa = 9.59RR75 pKa = 11.84VAAKK79 pKa = 10.12RR80 pKa = 11.84ATSKK84 pKa = 9.36TRR86 pKa = 11.84ASRR89 pKa = 3.35
Molecular weight: 9.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2550868 |
26 |
5084 |
323.4 |
34.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.444 ± 0.047 | 0.932 ± 0.01 |
5.696 ± 0.026 | 4.838 ± 0.026 |
3.477 ± 0.017 | 8.82 ± 0.035 |
2.138 ± 0.014 | 4.731 ± 0.02 |
2.825 ± 0.021 | 10.112 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.474 ± 0.013 | 2.253 ± 0.015 |
5.682 ± 0.026 | 3.145 ± 0.013 |
7.931 ± 0.033 | 4.841 ± 0.021 |
5.214 ± 0.029 | 7.72 ± 0.028 |
1.513 ± 0.013 | 2.215 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
