
Oleibacter marinus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Oleibacter
Average proteome isoelectric point is 5.74
Get precalculated fractions of proteins
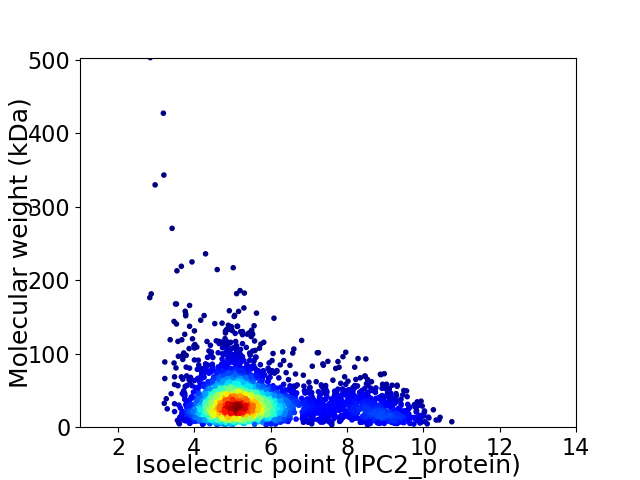
Virtual 2D-PAGE plot for 3470 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N7N1X4|A0A1N7N1X4_9GAMM Predicted membrane protein OS=Oleibacter marinus OX=484498 GN=SAMN05421686_106113 PE=4 SV=1
MM1 pKa = 6.31TTSLFSRR8 pKa = 11.84ASLLAITVSLMACGGGSGGSSNNDD32 pKa = 3.23QSPDD36 pKa = 3.47SADD39 pKa = 3.31NSTDD43 pKa = 3.46NPSGGDD49 pKa = 3.44QNTDD53 pKa = 2.99NTDD56 pKa = 3.34TQIVLSDD63 pKa = 3.85TTWSAGNDD71 pKa = 3.6RR72 pKa = 11.84ILGMGFNGDD81 pKa = 3.81SEE83 pKa = 5.0SPEE86 pKa = 3.84LTIMKK91 pKa = 8.34GAPVSGQDD99 pKa = 3.18GDD101 pKa = 3.68IFSIRR106 pKa = 11.84SVTYY110 pKa = 9.23PSTEE114 pKa = 3.56LSEE117 pKa = 4.25IKK119 pKa = 10.75SIDD122 pKa = 3.47NQADD126 pKa = 3.49RR127 pKa = 11.84RR128 pKa = 11.84DD129 pKa = 3.93FQDD132 pKa = 3.05MDD134 pKa = 3.33IVSYY138 pKa = 11.13NDD140 pKa = 2.94EE141 pKa = 4.67SYY143 pKa = 11.56VVACQDD149 pKa = 3.39GSDD152 pKa = 3.78TPGYY156 pKa = 8.19TSAVSLHH163 pKa = 6.44IYY165 pKa = 7.77KK166 pKa = 10.26TSDD169 pKa = 2.82TSKK172 pKa = 10.31VAEE175 pKa = 4.28IDD177 pKa = 3.89LVDD180 pKa = 4.1GSVEE184 pKa = 3.7ARR186 pKa = 11.84EE187 pKa = 4.28CSGIAAEE194 pKa = 4.65FTQGTSDD201 pKa = 3.79DD202 pKa = 3.96MGAVVYY208 pKa = 9.79YY209 pKa = 10.3AGNAMTTSGSPQYY222 pKa = 10.71SYY224 pKa = 11.62SRR226 pKa = 11.84IMKK229 pKa = 9.89VEE231 pKa = 3.73LTIDD235 pKa = 3.09TTANVDD241 pKa = 3.11ATDD244 pKa = 3.87AVVLDD249 pKa = 5.29DD250 pKa = 3.5ISTVVRR256 pKa = 11.84ASQTEE261 pKa = 4.04DD262 pKa = 3.02FMSGVTGFGDD272 pKa = 3.41HH273 pKa = 6.82VYY275 pKa = 8.94YY276 pKa = 10.32TYY278 pKa = 11.5YY279 pKa = 10.81DD280 pKa = 3.89DD281 pKa = 6.13SEE283 pKa = 4.33EE284 pKa = 4.45TNNLMYY290 pKa = 10.67ANSGSSPATIRR301 pKa = 11.84NVDD304 pKa = 3.09WSANDD309 pKa = 3.68PFVTGVAQQMLVKK322 pKa = 10.65DD323 pKa = 3.63MFVIPGQQATDD334 pKa = 3.57FDD336 pKa = 5.03SIYY339 pKa = 10.07MVSDD343 pKa = 3.9SAASGIVVAGYY354 pKa = 10.14RR355 pKa = 11.84RR356 pKa = 11.84DD357 pKa = 3.69MNLKK361 pKa = 9.65QAMTLSSDD369 pKa = 3.57TSVQNCSDD377 pKa = 4.39VITGISDD384 pKa = 3.09QGVGQKK390 pKa = 9.53LWCHH394 pKa = 6.64DD395 pKa = 3.56ATDD398 pKa = 3.6SGNIIEE404 pKa = 5.02VYY406 pKa = 10.77SPVHH410 pKa = 6.2PGNN413 pKa = 3.48
MM1 pKa = 6.31TTSLFSRR8 pKa = 11.84ASLLAITVSLMACGGGSGGSSNNDD32 pKa = 3.23QSPDD36 pKa = 3.47SADD39 pKa = 3.31NSTDD43 pKa = 3.46NPSGGDD49 pKa = 3.44QNTDD53 pKa = 2.99NTDD56 pKa = 3.34TQIVLSDD63 pKa = 3.85TTWSAGNDD71 pKa = 3.6RR72 pKa = 11.84ILGMGFNGDD81 pKa = 3.81SEE83 pKa = 5.0SPEE86 pKa = 3.84LTIMKK91 pKa = 8.34GAPVSGQDD99 pKa = 3.18GDD101 pKa = 3.68IFSIRR106 pKa = 11.84SVTYY110 pKa = 9.23PSTEE114 pKa = 3.56LSEE117 pKa = 4.25IKK119 pKa = 10.75SIDD122 pKa = 3.47NQADD126 pKa = 3.49RR127 pKa = 11.84RR128 pKa = 11.84DD129 pKa = 3.93FQDD132 pKa = 3.05MDD134 pKa = 3.33IVSYY138 pKa = 11.13NDD140 pKa = 2.94EE141 pKa = 4.67SYY143 pKa = 11.56VVACQDD149 pKa = 3.39GSDD152 pKa = 3.78TPGYY156 pKa = 8.19TSAVSLHH163 pKa = 6.44IYY165 pKa = 7.77KK166 pKa = 10.26TSDD169 pKa = 2.82TSKK172 pKa = 10.31VAEE175 pKa = 4.28IDD177 pKa = 3.89LVDD180 pKa = 4.1GSVEE184 pKa = 3.7ARR186 pKa = 11.84EE187 pKa = 4.28CSGIAAEE194 pKa = 4.65FTQGTSDD201 pKa = 3.79DD202 pKa = 3.96MGAVVYY208 pKa = 9.79YY209 pKa = 10.3AGNAMTTSGSPQYY222 pKa = 10.71SYY224 pKa = 11.62SRR226 pKa = 11.84IMKK229 pKa = 9.89VEE231 pKa = 3.73LTIDD235 pKa = 3.09TTANVDD241 pKa = 3.11ATDD244 pKa = 3.87AVVLDD249 pKa = 5.29DD250 pKa = 3.5ISTVVRR256 pKa = 11.84ASQTEE261 pKa = 4.04DD262 pKa = 3.02FMSGVTGFGDD272 pKa = 3.41HH273 pKa = 6.82VYY275 pKa = 8.94YY276 pKa = 10.32TYY278 pKa = 11.5YY279 pKa = 10.81DD280 pKa = 3.89DD281 pKa = 6.13SEE283 pKa = 4.33EE284 pKa = 4.45TNNLMYY290 pKa = 10.67ANSGSSPATIRR301 pKa = 11.84NVDD304 pKa = 3.09WSANDD309 pKa = 3.68PFVTGVAQQMLVKK322 pKa = 10.65DD323 pKa = 3.63MFVIPGQQATDD334 pKa = 3.57FDD336 pKa = 5.03SIYY339 pKa = 10.07MVSDD343 pKa = 3.9SAASGIVVAGYY354 pKa = 10.14RR355 pKa = 11.84RR356 pKa = 11.84DD357 pKa = 3.69MNLKK361 pKa = 9.65QAMTLSSDD369 pKa = 3.57TSVQNCSDD377 pKa = 4.39VITGISDD384 pKa = 3.09QGVGQKK390 pKa = 9.53LWCHH394 pKa = 6.64DD395 pKa = 3.56ATDD398 pKa = 3.6SGNIIEE404 pKa = 5.02VYY406 pKa = 10.77SPVHH410 pKa = 6.2PGNN413 pKa = 3.48
Molecular weight: 43.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N7Q192|A0A1N7Q192_9GAMM Uncharacterized protein OS=Oleibacter marinus OX=484498 GN=SAMN05421686_11290 PE=4 SV=1
MM1 pKa = 7.65DD2 pKa = 4.08TQRR5 pKa = 11.84LMALEE10 pKa = 4.53GSWLEE15 pKa = 3.85EE16 pKa = 3.83RR17 pKa = 11.84VSHH20 pKa = 6.49LRR22 pKa = 11.84GRR24 pKa = 11.84HH25 pKa = 5.08LLYY28 pKa = 10.88AGVDD32 pKa = 3.61TDD34 pKa = 4.01PKK36 pKa = 10.44FLGRR40 pKa = 11.84SRR42 pKa = 11.84ADD44 pKa = 2.91HH45 pKa = 5.93TFRR48 pKa = 11.84VGLPWQKK55 pKa = 11.3GLVDD59 pKa = 4.2CQAQIDD65 pKa = 3.94NTRR68 pKa = 11.84WPFPDD73 pKa = 3.08EE74 pKa = 4.07TLDD77 pKa = 4.0VVVLQHH83 pKa = 6.7ALDD86 pKa = 3.84MTSRR90 pKa = 11.84PHH92 pKa = 5.66QLVRR96 pKa = 11.84EE97 pKa = 4.44ATRR100 pKa = 11.84SLVSGGYY107 pKa = 9.87LVVVGFNPYY116 pKa = 8.36STWGGVRR123 pKa = 11.84WLRR126 pKa = 11.84TMSTEE131 pKa = 4.33LPWVSNPVAPLRR143 pKa = 11.84LSDD146 pKa = 3.51WLMLLDD152 pKa = 4.61FRR154 pKa = 11.84VEE156 pKa = 4.42NITTAAHH163 pKa = 6.6LWPVKK168 pKa = 10.1IGSEE172 pKa = 3.96AMSRR176 pKa = 11.84RR177 pKa = 11.84VDD179 pKa = 3.25RR180 pKa = 11.84VLAGNRR186 pKa = 11.84MVAGNIYY193 pKa = 10.56LLVARR198 pKa = 11.84KK199 pKa = 7.18TVAGMTTIRR208 pKa = 11.84SGRR211 pKa = 11.84KK212 pKa = 6.98VRR214 pKa = 11.84RR215 pKa = 11.84EE216 pKa = 3.9PKK218 pKa = 10.22LGFAIPTTRR227 pKa = 11.84KK228 pKa = 8.16PVSRR232 pKa = 11.84IKK234 pKa = 11.02GSLL237 pKa = 3.22
MM1 pKa = 7.65DD2 pKa = 4.08TQRR5 pKa = 11.84LMALEE10 pKa = 4.53GSWLEE15 pKa = 3.85EE16 pKa = 3.83RR17 pKa = 11.84VSHH20 pKa = 6.49LRR22 pKa = 11.84GRR24 pKa = 11.84HH25 pKa = 5.08LLYY28 pKa = 10.88AGVDD32 pKa = 3.61TDD34 pKa = 4.01PKK36 pKa = 10.44FLGRR40 pKa = 11.84SRR42 pKa = 11.84ADD44 pKa = 2.91HH45 pKa = 5.93TFRR48 pKa = 11.84VGLPWQKK55 pKa = 11.3GLVDD59 pKa = 4.2CQAQIDD65 pKa = 3.94NTRR68 pKa = 11.84WPFPDD73 pKa = 3.08EE74 pKa = 4.07TLDD77 pKa = 4.0VVVLQHH83 pKa = 6.7ALDD86 pKa = 3.84MTSRR90 pKa = 11.84PHH92 pKa = 5.66QLVRR96 pKa = 11.84EE97 pKa = 4.44ATRR100 pKa = 11.84SLVSGGYY107 pKa = 9.87LVVVGFNPYY116 pKa = 8.36STWGGVRR123 pKa = 11.84WLRR126 pKa = 11.84TMSTEE131 pKa = 4.33LPWVSNPVAPLRR143 pKa = 11.84LSDD146 pKa = 3.51WLMLLDD152 pKa = 4.61FRR154 pKa = 11.84VEE156 pKa = 4.42NITTAAHH163 pKa = 6.6LWPVKK168 pKa = 10.1IGSEE172 pKa = 3.96AMSRR176 pKa = 11.84RR177 pKa = 11.84VDD179 pKa = 3.25RR180 pKa = 11.84VLAGNRR186 pKa = 11.84MVAGNIYY193 pKa = 10.56LLVARR198 pKa = 11.84KK199 pKa = 7.18TVAGMTTIRR208 pKa = 11.84SGRR211 pKa = 11.84KK212 pKa = 6.98VRR214 pKa = 11.84RR215 pKa = 11.84EE216 pKa = 3.9PKK218 pKa = 10.22LGFAIPTTRR227 pKa = 11.84KK228 pKa = 8.16PVSRR232 pKa = 11.84IKK234 pKa = 11.02GSLL237 pKa = 3.22
Molecular weight: 26.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1171308 |
27 |
5033 |
337.6 |
37.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.425 ± 0.053 | 0.932 ± 0.016 |
6.417 ± 0.079 | 6.788 ± 0.046 |
3.748 ± 0.028 | 7.299 ± 0.042 |
2.082 ± 0.022 | 5.593 ± 0.029 |
4.092 ± 0.046 | 10.136 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 0.024 | 3.752 ± 0.043 |
4.239 ± 0.034 | 3.968 ± 0.029 |
5.538 ± 0.047 | 6.729 ± 0.038 |
5.534 ± 0.06 | 7.021 ± 0.04 |
1.302 ± 0.017 | 2.863 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
