
Aeromicrobium sp. Leaf289
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Aeromicrobium; unclassified Aeromicrobium
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
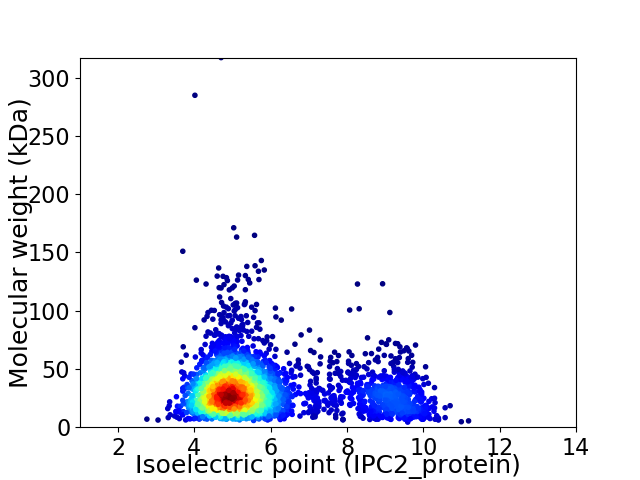
Virtual 2D-PAGE plot for 3243 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S9RD88|A0A0S9RD88_9ACTN N-acetyltransferase domain-containing protein OS=Aeromicrobium sp. Leaf289 OX=1736324 GN=ASF37_02760 PE=4 SV=1
MM1 pKa = 8.11IPMFLLATHH10 pKa = 6.81RR11 pKa = 11.84RR12 pKa = 11.84SEE14 pKa = 4.08RR15 pKa = 11.84GQGSLEE21 pKa = 4.16TVGVVVLASILVAATTGVVVQSSPALRR48 pKa = 11.84SEE50 pKa = 3.7VGYY53 pKa = 9.72RR54 pKa = 11.84VCQITSLGGGGCEE67 pKa = 4.94DD68 pKa = 4.9PGDD71 pKa = 4.92EE72 pKa = 5.25IPGDD76 pKa = 4.13DD77 pKa = 4.73GADD80 pKa = 3.48PDD82 pKa = 4.92SPWGNEE88 pKa = 3.56NARR91 pKa = 11.84PDD93 pKa = 3.67GKK95 pKa = 9.94PAVDD99 pKa = 3.88EE100 pKa = 4.35SRR102 pKa = 11.84LPDD105 pKa = 3.68EE106 pKa = 4.76PCVVSSTSGSISATGSFVVSATGEE130 pKa = 3.71QGMTIEE136 pKa = 4.36TLSDD140 pKa = 2.89GTYY143 pKa = 9.81RR144 pKa = 11.84VSVHH148 pKa = 6.9DD149 pKa = 5.09AGSLGYY155 pKa = 10.4GVGIGVDD162 pKa = 3.64GNVVVDD168 pKa = 4.85GVQQGSTAYY177 pKa = 10.1AGADD181 pKa = 2.93IALAGLASQTYY192 pKa = 8.64YY193 pKa = 11.0AGSEE197 pKa = 4.09QEE199 pKa = 4.29LQDD202 pKa = 3.78LLVRR206 pKa = 11.84VAGEE210 pKa = 3.69QALDD214 pKa = 3.53QVAPRR219 pKa = 11.84IPGPADD225 pKa = 2.91IPFVQDD231 pKa = 2.95IPVIGEE237 pKa = 4.37LEE239 pKa = 4.06ALDD242 pKa = 4.32NPVRR246 pKa = 11.84WAVDD250 pKa = 3.38QLAGPPPEE258 pKa = 5.0PDD260 pKa = 3.01EE261 pKa = 5.19QYY263 pKa = 11.32VQLGLEE269 pKa = 4.44GDD271 pKa = 3.84ASAALAGIVYY281 pKa = 8.55STGAEE286 pKa = 3.66LDD288 pKa = 3.38MGVFAGARR296 pKa = 11.84TTPEE300 pKa = 4.51GYY302 pKa = 10.44VLTSVVEE309 pKa = 4.46ANGSAWASAFGGEE322 pKa = 4.1AQAMAGGTLVRR333 pKa = 11.84EE334 pKa = 4.12IHH336 pKa = 6.65LGPDD340 pKa = 3.66GTPLGITMTLTGGANAQSGDD360 pKa = 3.74VGPATDD366 pKa = 3.63TQDD369 pKa = 3.43SYY371 pKa = 12.13VATAYY376 pKa = 10.79VPFTGDD382 pKa = 2.68VATDD386 pKa = 3.16AGIYY390 pKa = 10.16LSAANMYY397 pKa = 8.03QAPQFLQSAMEE408 pKa = 4.24TGTVSLDD415 pKa = 3.42QYY417 pKa = 11.18EE418 pKa = 4.48VDD420 pKa = 3.71PNTYY424 pKa = 9.26GANVSIDD431 pKa = 3.17IPGAEE436 pKa = 4.64GGLGVNGDD444 pKa = 4.11LTNSTMVSSSYY455 pKa = 10.0WDD457 pKa = 3.15GSGFVDD463 pKa = 5.01RR464 pKa = 11.84GCC466 pKa = 4.37
MM1 pKa = 8.11IPMFLLATHH10 pKa = 6.81RR11 pKa = 11.84RR12 pKa = 11.84SEE14 pKa = 4.08RR15 pKa = 11.84GQGSLEE21 pKa = 4.16TVGVVVLASILVAATTGVVVQSSPALRR48 pKa = 11.84SEE50 pKa = 3.7VGYY53 pKa = 9.72RR54 pKa = 11.84VCQITSLGGGGCEE67 pKa = 4.94DD68 pKa = 4.9PGDD71 pKa = 4.92EE72 pKa = 5.25IPGDD76 pKa = 4.13DD77 pKa = 4.73GADD80 pKa = 3.48PDD82 pKa = 4.92SPWGNEE88 pKa = 3.56NARR91 pKa = 11.84PDD93 pKa = 3.67GKK95 pKa = 9.94PAVDD99 pKa = 3.88EE100 pKa = 4.35SRR102 pKa = 11.84LPDD105 pKa = 3.68EE106 pKa = 4.76PCVVSSTSGSISATGSFVVSATGEE130 pKa = 3.71QGMTIEE136 pKa = 4.36TLSDD140 pKa = 2.89GTYY143 pKa = 9.81RR144 pKa = 11.84VSVHH148 pKa = 6.9DD149 pKa = 5.09AGSLGYY155 pKa = 10.4GVGIGVDD162 pKa = 3.64GNVVVDD168 pKa = 4.85GVQQGSTAYY177 pKa = 10.1AGADD181 pKa = 2.93IALAGLASQTYY192 pKa = 8.64YY193 pKa = 11.0AGSEE197 pKa = 4.09QEE199 pKa = 4.29LQDD202 pKa = 3.78LLVRR206 pKa = 11.84VAGEE210 pKa = 3.69QALDD214 pKa = 3.53QVAPRR219 pKa = 11.84IPGPADD225 pKa = 2.91IPFVQDD231 pKa = 2.95IPVIGEE237 pKa = 4.37LEE239 pKa = 4.06ALDD242 pKa = 4.32NPVRR246 pKa = 11.84WAVDD250 pKa = 3.38QLAGPPPEE258 pKa = 5.0PDD260 pKa = 3.01EE261 pKa = 5.19QYY263 pKa = 11.32VQLGLEE269 pKa = 4.44GDD271 pKa = 3.84ASAALAGIVYY281 pKa = 8.55STGAEE286 pKa = 3.66LDD288 pKa = 3.38MGVFAGARR296 pKa = 11.84TTPEE300 pKa = 4.51GYY302 pKa = 10.44VLTSVVEE309 pKa = 4.46ANGSAWASAFGGEE322 pKa = 4.1AQAMAGGTLVRR333 pKa = 11.84EE334 pKa = 4.12IHH336 pKa = 6.65LGPDD340 pKa = 3.66GTPLGITMTLTGGANAQSGDD360 pKa = 3.74VGPATDD366 pKa = 3.63TQDD369 pKa = 3.43SYY371 pKa = 12.13VATAYY376 pKa = 10.79VPFTGDD382 pKa = 2.68VATDD386 pKa = 3.16AGIYY390 pKa = 10.16LSAANMYY397 pKa = 8.03QAPQFLQSAMEE408 pKa = 4.24TGTVSLDD415 pKa = 3.42QYY417 pKa = 11.18EE418 pKa = 4.48VDD420 pKa = 3.71PNTYY424 pKa = 9.26GANVSIDD431 pKa = 3.17IPGAEE436 pKa = 4.64GGLGVNGDD444 pKa = 4.11LTNSTMVSSSYY455 pKa = 10.0WDD457 pKa = 3.15GSGFVDD463 pKa = 5.01RR464 pKa = 11.84GCC466 pKa = 4.37
Molecular weight: 47.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S9R810|A0A0S9R810_9ACTN Uncharacterized protein OS=Aeromicrobium sp. Leaf289 OX=1736324 GN=ASF37_06645 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.83GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.83GRR40 pKa = 11.84AKK42 pKa = 10.68LSAA45 pKa = 3.92
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1030146 |
37 |
3044 |
317.7 |
33.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.962 ± 0.052 | 0.653 ± 0.01 |
6.91 ± 0.039 | 5.761 ± 0.041 |
2.769 ± 0.025 | 9.163 ± 0.039 |
2.172 ± 0.022 | 3.219 ± 0.032 |
1.935 ± 0.033 | 10.21 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.695 ± 0.018 | 1.609 ± 0.022 |
5.41 ± 0.032 | 2.74 ± 0.022 |
7.672 ± 0.051 | 5.426 ± 0.031 |
6.224 ± 0.041 | 10.154 ± 0.046 |
1.471 ± 0.021 | 1.845 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
