
Muricauda pacifica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Muricauda
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
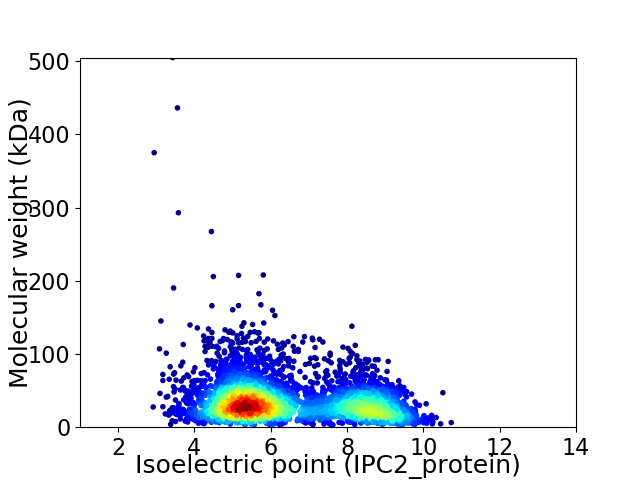
Virtual 2D-PAGE plot for 4085 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T0M905|A0A2T0M905_9FLAO CubicO group peptidase (Beta-lactamase class C family) OS=Muricauda pacifica OX=1080225 GN=CLV81_2352 PE=4 SV=1
MM1 pKa = 7.44EE2 pKa = 4.84MSAQNSGDD10 pKa = 3.74CRR12 pKa = 11.84TAIPVCADD20 pKa = 3.43MPILSATDD28 pKa = 3.77GSGDD32 pKa = 3.86IDD34 pKa = 5.58DD35 pKa = 5.54FDD37 pKa = 6.76PDD39 pKa = 4.14VITQTGCLEE48 pKa = 4.3KK49 pKa = 11.05GSVASANIEE58 pKa = 4.33NNTSWYY64 pKa = 9.72VFRR67 pKa = 11.84AGTDD71 pKa = 3.48GQIGFDD77 pKa = 3.57IEE79 pKa = 4.38ALPVTPGGIVTSEE92 pKa = 3.8IDD94 pKa = 3.28FALYY98 pKa = 10.7GPFDD102 pKa = 3.51QTLGSNFCTTIGNGSDD118 pKa = 2.81IPIRR122 pKa = 11.84CNYY125 pKa = 8.87EE126 pKa = 3.57INDD129 pKa = 3.83TGFTGIGVNPISGQVGAPFVRR150 pKa = 11.84ASQNTYY156 pKa = 10.99DD157 pKa = 3.49EE158 pKa = 4.42WLDD161 pKa = 3.62VQQGEE166 pKa = 4.4IYY168 pKa = 10.92YY169 pKa = 10.54LFINNFNTNFNGDD182 pKa = 4.1PEE184 pKa = 4.56PFSLTFTGSSVDD196 pKa = 3.44ADD198 pKa = 3.46QNTALDD204 pKa = 3.73CTLRR208 pKa = 11.84DD209 pKa = 3.55EE210 pKa = 5.33FLGLDD215 pKa = 4.04IIACEE220 pKa = 4.1GDD222 pKa = 3.41PDD224 pKa = 4.1IVLSALNSPAGPNIANITWEE244 pKa = 4.17VDD246 pKa = 3.43LEE248 pKa = 4.43DD249 pKa = 5.42DD250 pKa = 4.2GVYY253 pKa = 10.77DD254 pKa = 4.03QVLATGAGATEE265 pKa = 4.29LTVTSPNSGRR275 pKa = 11.84YY276 pKa = 8.68RR277 pKa = 11.84VTIEE281 pKa = 3.88TTFATIITDD290 pKa = 4.41DD291 pKa = 3.32ILITFFGVPVLDD303 pKa = 3.92EE304 pKa = 4.23VRR306 pKa = 11.84VLDD309 pKa = 4.85DD310 pKa = 5.17LVNSDD315 pKa = 3.35QTDD318 pKa = 3.73PYY320 pKa = 10.47NIEE323 pKa = 3.73ISVLGNGNYY332 pKa = 9.99EE333 pKa = 3.88YY334 pKa = 10.41AINGGEE340 pKa = 4.09FQDD343 pKa = 4.94DD344 pKa = 4.14PVFLDD349 pKa = 3.73VPPGVNTVVINDD361 pKa = 3.43VNGCGTTEE369 pKa = 4.51PIEE372 pKa = 4.07FLVVGYY378 pKa = 9.34PKK380 pKa = 10.71FFTPNGDD387 pKa = 3.92GIHH390 pKa = 7.23DD391 pKa = 3.74NWNVLGVEE399 pKa = 4.3TLTNPSIFIFDD410 pKa = 3.74RR411 pKa = 11.84YY412 pKa = 11.07GKK414 pKa = 9.85FLKK417 pKa = 10.63QLDD420 pKa = 4.08INSLGWDD427 pKa = 3.44GTFNGRR433 pKa = 11.84PLPSSDD439 pKa = 2.51YY440 pKa = 10.38WFRR443 pKa = 11.84LDD445 pKa = 4.67YY446 pKa = 11.16EE447 pKa = 4.66RR448 pKa = 11.84DD449 pKa = 3.65DD450 pKa = 4.16EE451 pKa = 4.65GVLVARR457 pKa = 11.84SVRR460 pKa = 11.84RR461 pKa = 11.84HH462 pKa = 5.32FSLIRR467 pKa = 4.08
MM1 pKa = 7.44EE2 pKa = 4.84MSAQNSGDD10 pKa = 3.74CRR12 pKa = 11.84TAIPVCADD20 pKa = 3.43MPILSATDD28 pKa = 3.77GSGDD32 pKa = 3.86IDD34 pKa = 5.58DD35 pKa = 5.54FDD37 pKa = 6.76PDD39 pKa = 4.14VITQTGCLEE48 pKa = 4.3KK49 pKa = 11.05GSVASANIEE58 pKa = 4.33NNTSWYY64 pKa = 9.72VFRR67 pKa = 11.84AGTDD71 pKa = 3.48GQIGFDD77 pKa = 3.57IEE79 pKa = 4.38ALPVTPGGIVTSEE92 pKa = 3.8IDD94 pKa = 3.28FALYY98 pKa = 10.7GPFDD102 pKa = 3.51QTLGSNFCTTIGNGSDD118 pKa = 2.81IPIRR122 pKa = 11.84CNYY125 pKa = 8.87EE126 pKa = 3.57INDD129 pKa = 3.83TGFTGIGVNPISGQVGAPFVRR150 pKa = 11.84ASQNTYY156 pKa = 10.99DD157 pKa = 3.49EE158 pKa = 4.42WLDD161 pKa = 3.62VQQGEE166 pKa = 4.4IYY168 pKa = 10.92YY169 pKa = 10.54LFINNFNTNFNGDD182 pKa = 4.1PEE184 pKa = 4.56PFSLTFTGSSVDD196 pKa = 3.44ADD198 pKa = 3.46QNTALDD204 pKa = 3.73CTLRR208 pKa = 11.84DD209 pKa = 3.55EE210 pKa = 5.33FLGLDD215 pKa = 4.04IIACEE220 pKa = 4.1GDD222 pKa = 3.41PDD224 pKa = 4.1IVLSALNSPAGPNIANITWEE244 pKa = 4.17VDD246 pKa = 3.43LEE248 pKa = 4.43DD249 pKa = 5.42DD250 pKa = 4.2GVYY253 pKa = 10.77DD254 pKa = 4.03QVLATGAGATEE265 pKa = 4.29LTVTSPNSGRR275 pKa = 11.84YY276 pKa = 8.68RR277 pKa = 11.84VTIEE281 pKa = 3.88TTFATIITDD290 pKa = 4.41DD291 pKa = 3.32ILITFFGVPVLDD303 pKa = 3.92EE304 pKa = 4.23VRR306 pKa = 11.84VLDD309 pKa = 4.85DD310 pKa = 5.17LVNSDD315 pKa = 3.35QTDD318 pKa = 3.73PYY320 pKa = 10.47NIEE323 pKa = 3.73ISVLGNGNYY332 pKa = 9.99EE333 pKa = 3.88YY334 pKa = 10.41AINGGEE340 pKa = 4.09FQDD343 pKa = 4.94DD344 pKa = 4.14PVFLDD349 pKa = 3.73VPPGVNTVVINDD361 pKa = 3.43VNGCGTTEE369 pKa = 4.51PIEE372 pKa = 4.07FLVVGYY378 pKa = 9.34PKK380 pKa = 10.71FFTPNGDD387 pKa = 3.92GIHH390 pKa = 7.23DD391 pKa = 3.74NWNVLGVEE399 pKa = 4.3TLTNPSIFIFDD410 pKa = 3.74RR411 pKa = 11.84YY412 pKa = 11.07GKK414 pKa = 9.85FLKK417 pKa = 10.63QLDD420 pKa = 4.08INSLGWDD427 pKa = 3.44GTFNGRR433 pKa = 11.84PLPSSDD439 pKa = 2.51YY440 pKa = 10.38WFRR443 pKa = 11.84LDD445 pKa = 4.67YY446 pKa = 11.16EE447 pKa = 4.66RR448 pKa = 11.84DD449 pKa = 3.65DD450 pKa = 4.16EE451 pKa = 4.65GVLVARR457 pKa = 11.84SVRR460 pKa = 11.84RR461 pKa = 11.84HH462 pKa = 5.32FSLIRR467 pKa = 4.08
Molecular weight: 50.84 kDa
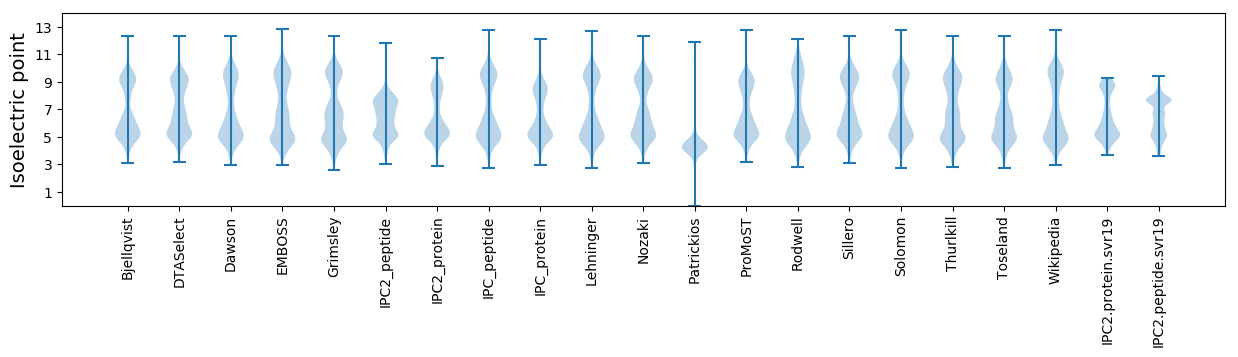
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T0MJ46|A0A2T0MJ46_9FLAO Uncharacterized protein OS=Muricauda pacifica OX=1080225 GN=CLV81_1581 PE=4 SV=1
MM1 pKa = 7.07TLQRR5 pKa = 11.84FRR7 pKa = 11.84ITFSGEE13 pKa = 3.68ANKK16 pKa = 9.72PLPIQIKK23 pKa = 8.91PNVVKK28 pKa = 10.74ILGRR32 pKa = 11.84LGLYY36 pKa = 9.91KK37 pKa = 10.37ISPLRR42 pKa = 11.84IYY44 pKa = 10.97VVGCPFLFVV53 pKa = 3.5
MM1 pKa = 7.07TLQRR5 pKa = 11.84FRR7 pKa = 11.84ITFSGEE13 pKa = 3.68ANKK16 pKa = 9.72PLPIQIKK23 pKa = 8.91PNVVKK28 pKa = 10.74ILGRR32 pKa = 11.84LGLYY36 pKa = 9.91KK37 pKa = 10.37ISPLRR42 pKa = 11.84IYY44 pKa = 10.97VVGCPFLFVV53 pKa = 3.5
Molecular weight: 6.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1342224 |
29 |
4767 |
328.6 |
37.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.214 ± 0.035 | 0.722 ± 0.011 |
5.631 ± 0.035 | 6.94 ± 0.035 |
5.404 ± 0.033 | 6.717 ± 0.036 |
1.826 ± 0.02 | 7.32 ± 0.032 |
7.189 ± 0.056 | 9.693 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.254 ± 0.02 | 5.648 ± 0.036 |
3.577 ± 0.023 | 3.484 ± 0.022 |
3.891 ± 0.029 | 6.559 ± 0.03 |
5.557 ± 0.049 | 6.38 ± 0.03 |
1.149 ± 0.015 | 3.844 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
