
Cybaeus spider associated circular virus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus cybusi1
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins
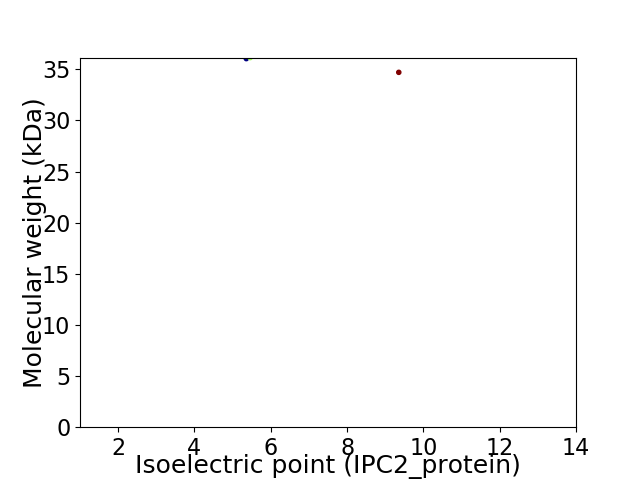
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
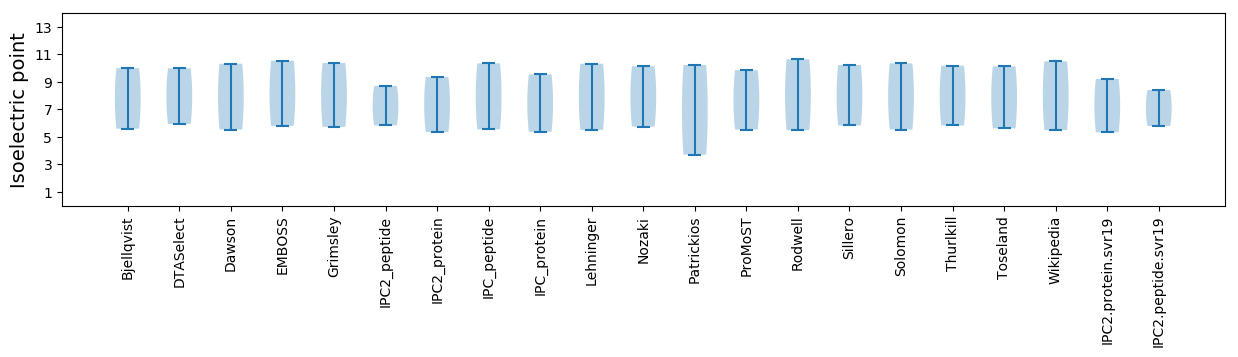
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BP79|A0A346BP79_9VIRU Putative capsid protein OS=Cybaeus spider associated circular virus 2 OX=2293278 PE=4 SV=1
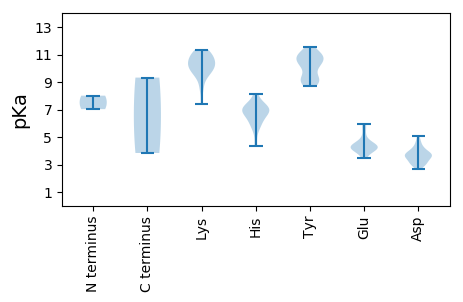
MM1 pKa = 8.01PFRR4 pKa = 11.84FQAKK8 pKa = 9.96YY9 pKa = 11.08GLLTYY14 pKa = 9.02AQCGDD19 pKa = 3.76LDD21 pKa = 3.69AFAIVDD27 pKa = 3.78HH28 pKa = 6.67LSEE31 pKa = 5.05LGAEE35 pKa = 4.44CIVGEE40 pKa = 4.46EE41 pKa = 3.98LHH43 pKa = 7.03ADD45 pKa = 4.18GGRR48 pKa = 11.84HH49 pKa = 4.36LHH51 pKa = 6.16AFFMFQHH58 pKa = 7.09KK59 pKa = 9.71FRR61 pKa = 11.84TSNARR66 pKa = 11.84VFDD69 pKa = 3.52VGGNHH74 pKa = 7.03ANISPGYY81 pKa = 9.2SNPSDD86 pKa = 3.89GYY88 pKa = 11.29DD89 pKa = 3.44YY90 pKa = 10.99AIKK93 pKa = 10.53DD94 pKa = 3.79GNILAGGLEE103 pKa = 4.2RR104 pKa = 11.84PGNEE108 pKa = 4.36SPKK111 pKa = 10.71DD112 pKa = 3.44DD113 pKa = 4.64SKK115 pKa = 11.12WHH117 pKa = 6.66TITAAEE123 pKa = 4.32TEE125 pKa = 4.51SEE127 pKa = 4.36FWALVAQLDD136 pKa = 3.99PRR138 pKa = 11.84SMVVSYY144 pKa = 9.43PALRR148 pKa = 11.84AYY150 pKa = 10.2AGNRR154 pKa = 11.84YY155 pKa = 9.13RR156 pKa = 11.84VDD158 pKa = 4.62RR159 pKa = 11.84IPYY162 pKa = 9.32RR163 pKa = 11.84NPDD166 pKa = 3.31GLRR169 pKa = 11.84YY170 pKa = 8.73CTDD173 pKa = 3.85EE174 pKa = 4.22YY175 pKa = 10.83PHH177 pKa = 8.16LDD179 pKa = 3.07EE180 pKa = 4.49WVEE183 pKa = 3.9GSLRR187 pKa = 11.84NNRR190 pKa = 11.84EE191 pKa = 3.47NRR193 pKa = 11.84PRR195 pKa = 11.84SLVLYY200 pKa = 10.49GGTRR204 pKa = 11.84LGKK207 pKa = 7.42TLWSRR212 pKa = 11.84SLGNHH217 pKa = 7.02AYY219 pKa = 10.42FGGLFAMDD227 pKa = 4.37EE228 pKa = 4.81PIDD231 pKa = 4.2DD232 pKa = 2.98VDD234 pKa = 3.84YY235 pKa = 11.56AVFDD239 pKa = 4.27DD240 pKa = 4.21MQGGLDD246 pKa = 3.81FFHH249 pKa = 7.45SYY251 pKa = 10.7KK252 pKa = 10.46FWLGAQKK259 pKa = 10.36QFYY262 pKa = 8.76VTDD265 pKa = 3.67KK266 pKa = 11.21YY267 pKa = 10.97KK268 pKa = 10.96GKK270 pKa = 10.72KK271 pKa = 8.43LVSWGKK277 pKa = 8.75PCIYY281 pKa = 10.26LSNNDD286 pKa = 3.16PRR288 pKa = 11.84GDD290 pKa = 3.57KK291 pKa = 10.95SCDD294 pKa = 3.41IEE296 pKa = 4.22WLEE299 pKa = 4.29GNCTIVYY306 pKa = 9.08IDD308 pKa = 3.88RR309 pKa = 11.84PIFRR313 pKa = 11.84ANTEE317 pKa = 3.84
MM1 pKa = 8.01PFRR4 pKa = 11.84FQAKK8 pKa = 9.96YY9 pKa = 11.08GLLTYY14 pKa = 9.02AQCGDD19 pKa = 3.76LDD21 pKa = 3.69AFAIVDD27 pKa = 3.78HH28 pKa = 6.67LSEE31 pKa = 5.05LGAEE35 pKa = 4.44CIVGEE40 pKa = 4.46EE41 pKa = 3.98LHH43 pKa = 7.03ADD45 pKa = 4.18GGRR48 pKa = 11.84HH49 pKa = 4.36LHH51 pKa = 6.16AFFMFQHH58 pKa = 7.09KK59 pKa = 9.71FRR61 pKa = 11.84TSNARR66 pKa = 11.84VFDD69 pKa = 3.52VGGNHH74 pKa = 7.03ANISPGYY81 pKa = 9.2SNPSDD86 pKa = 3.89GYY88 pKa = 11.29DD89 pKa = 3.44YY90 pKa = 10.99AIKK93 pKa = 10.53DD94 pKa = 3.79GNILAGGLEE103 pKa = 4.2RR104 pKa = 11.84PGNEE108 pKa = 4.36SPKK111 pKa = 10.71DD112 pKa = 3.44DD113 pKa = 4.64SKK115 pKa = 11.12WHH117 pKa = 6.66TITAAEE123 pKa = 4.32TEE125 pKa = 4.51SEE127 pKa = 4.36FWALVAQLDD136 pKa = 3.99PRR138 pKa = 11.84SMVVSYY144 pKa = 9.43PALRR148 pKa = 11.84AYY150 pKa = 10.2AGNRR154 pKa = 11.84YY155 pKa = 9.13RR156 pKa = 11.84VDD158 pKa = 4.62RR159 pKa = 11.84IPYY162 pKa = 9.32RR163 pKa = 11.84NPDD166 pKa = 3.31GLRR169 pKa = 11.84YY170 pKa = 8.73CTDD173 pKa = 3.85EE174 pKa = 4.22YY175 pKa = 10.83PHH177 pKa = 8.16LDD179 pKa = 3.07EE180 pKa = 4.49WVEE183 pKa = 3.9GSLRR187 pKa = 11.84NNRR190 pKa = 11.84EE191 pKa = 3.47NRR193 pKa = 11.84PRR195 pKa = 11.84SLVLYY200 pKa = 10.49GGTRR204 pKa = 11.84LGKK207 pKa = 7.42TLWSRR212 pKa = 11.84SLGNHH217 pKa = 7.02AYY219 pKa = 10.42FGGLFAMDD227 pKa = 4.37EE228 pKa = 4.81PIDD231 pKa = 4.2DD232 pKa = 2.98VDD234 pKa = 3.84YY235 pKa = 11.56AVFDD239 pKa = 4.27DD240 pKa = 4.21MQGGLDD246 pKa = 3.81FFHH249 pKa = 7.45SYY251 pKa = 10.7KK252 pKa = 10.46FWLGAQKK259 pKa = 10.36QFYY262 pKa = 8.76VTDD265 pKa = 3.67KK266 pKa = 11.21YY267 pKa = 10.97KK268 pKa = 10.96GKK270 pKa = 10.72KK271 pKa = 8.43LVSWGKK277 pKa = 8.75PCIYY281 pKa = 10.26LSNNDD286 pKa = 3.16PRR288 pKa = 11.84GDD290 pKa = 3.57KK291 pKa = 10.95SCDD294 pKa = 3.41IEE296 pKa = 4.22WLEE299 pKa = 4.29GNCTIVYY306 pKa = 9.08IDD308 pKa = 3.88RR309 pKa = 11.84PIFRR313 pKa = 11.84ANTEE317 pKa = 3.84
Molecular weight: 36.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BP79|A0A346BP79_9VIRU Putative capsid protein OS=Cybaeus spider associated circular virus 2 OX=2293278 PE=4 SV=1
MM1 pKa = 7.03VFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84STLRR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84PARR17 pKa = 11.84RR18 pKa = 11.84TRR20 pKa = 11.84RR21 pKa = 11.84SNPRR25 pKa = 11.84RR26 pKa = 11.84SYY28 pKa = 10.44AKK30 pKa = 9.36KK31 pKa = 9.88RR32 pKa = 11.84SFKK35 pKa = 10.66RR36 pKa = 11.84RR37 pKa = 11.84TTTRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84FPSTKK48 pKa = 9.6MILNKK53 pKa = 10.08TSTKK57 pKa = 10.29KK58 pKa = 9.55RR59 pKa = 11.84DD60 pKa = 3.01KK61 pKa = 10.13MLPHH65 pKa = 6.95GSANTDD71 pKa = 2.75ADD73 pKa = 3.26IAGGVTISSGDD84 pKa = 3.53GYY86 pKa = 9.9FTSGWIATARR96 pKa = 11.84DD97 pKa = 3.4KK98 pKa = 11.26FKK100 pKa = 11.35NDD102 pKa = 2.69GSLGTKK108 pKa = 10.34AEE110 pKa = 4.17MAVRR114 pKa = 11.84TAQTCFMVGLRR125 pKa = 11.84EE126 pKa = 3.96NVYY129 pKa = 10.84LRR131 pKa = 11.84TDD133 pKa = 5.09DD134 pKa = 3.89GASWQWRR141 pKa = 11.84RR142 pKa = 11.84ICFTFKK148 pKa = 10.75GSGIINNDD156 pKa = 3.36SQDD159 pKa = 3.52SSDD162 pKa = 3.71SGWFRR167 pKa = 11.84EE168 pKa = 4.1TSNGFVRR175 pKa = 11.84YY176 pKa = 9.16IHH178 pKa = 6.11QVPLGNNLLQVLFKK192 pKa = 11.04GALNVDD198 pKa = 2.98WTDD201 pKa = 3.13QFTAPVDD208 pKa = 3.68TTRR211 pKa = 11.84VSLKK215 pKa = 10.04YY216 pKa = 10.31DD217 pKa = 2.97KK218 pKa = 10.05QTIIRR223 pKa = 11.84SGNEE227 pKa = 3.45EE228 pKa = 4.06GVGRR232 pKa = 11.84MHH234 pKa = 6.92KK235 pKa = 9.39RR236 pKa = 11.84WHH238 pKa = 5.58GMYY241 pKa = 10.29HH242 pKa = 5.9NLVYY246 pKa = 10.89DD247 pKa = 3.7EE248 pKa = 5.82DD249 pKa = 4.79EE250 pKa = 5.48IGDD253 pKa = 4.1SMTDD257 pKa = 3.04NNLSVEE263 pKa = 4.56AKK265 pKa = 10.12PGMGDD270 pKa = 3.56YY271 pKa = 10.97YY272 pKa = 10.87VIDD275 pKa = 3.66MFRR278 pKa = 11.84CQSSGTDD285 pKa = 3.05HH286 pKa = 7.1EE287 pKa = 5.96LFFQPEE293 pKa = 4.22STLYY297 pKa = 9.05WHH299 pKa = 7.11EE300 pKa = 4.13KK301 pKa = 9.32
MM1 pKa = 7.03VFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84STLRR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84PARR17 pKa = 11.84RR18 pKa = 11.84TRR20 pKa = 11.84RR21 pKa = 11.84SNPRR25 pKa = 11.84RR26 pKa = 11.84SYY28 pKa = 10.44AKK30 pKa = 9.36KK31 pKa = 9.88RR32 pKa = 11.84SFKK35 pKa = 10.66RR36 pKa = 11.84RR37 pKa = 11.84TTTRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84FPSTKK48 pKa = 9.6MILNKK53 pKa = 10.08TSTKK57 pKa = 10.29KK58 pKa = 9.55RR59 pKa = 11.84DD60 pKa = 3.01KK61 pKa = 10.13MLPHH65 pKa = 6.95GSANTDD71 pKa = 2.75ADD73 pKa = 3.26IAGGVTISSGDD84 pKa = 3.53GYY86 pKa = 9.9FTSGWIATARR96 pKa = 11.84DD97 pKa = 3.4KK98 pKa = 11.26FKK100 pKa = 11.35NDD102 pKa = 2.69GSLGTKK108 pKa = 10.34AEE110 pKa = 4.17MAVRR114 pKa = 11.84TAQTCFMVGLRR125 pKa = 11.84EE126 pKa = 3.96NVYY129 pKa = 10.84LRR131 pKa = 11.84TDD133 pKa = 5.09DD134 pKa = 3.89GASWQWRR141 pKa = 11.84RR142 pKa = 11.84ICFTFKK148 pKa = 10.75GSGIINNDD156 pKa = 3.36SQDD159 pKa = 3.52SSDD162 pKa = 3.71SGWFRR167 pKa = 11.84EE168 pKa = 4.1TSNGFVRR175 pKa = 11.84YY176 pKa = 9.16IHH178 pKa = 6.11QVPLGNNLLQVLFKK192 pKa = 11.04GALNVDD198 pKa = 2.98WTDD201 pKa = 3.13QFTAPVDD208 pKa = 3.68TTRR211 pKa = 11.84VSLKK215 pKa = 10.04YY216 pKa = 10.31DD217 pKa = 2.97KK218 pKa = 10.05QTIIRR223 pKa = 11.84SGNEE227 pKa = 3.45EE228 pKa = 4.06GVGRR232 pKa = 11.84MHH234 pKa = 6.92KK235 pKa = 9.39RR236 pKa = 11.84WHH238 pKa = 5.58GMYY241 pKa = 10.29HH242 pKa = 5.9NLVYY246 pKa = 10.89DD247 pKa = 3.7EE248 pKa = 5.82DD249 pKa = 4.79EE250 pKa = 5.48IGDD253 pKa = 4.1SMTDD257 pKa = 3.04NNLSVEE263 pKa = 4.56AKK265 pKa = 10.12PGMGDD270 pKa = 3.56YY271 pKa = 10.97YY272 pKa = 10.87VIDD275 pKa = 3.66MFRR278 pKa = 11.84CQSSGTDD285 pKa = 3.05HH286 pKa = 7.1EE287 pKa = 5.96LFFQPEE293 pKa = 4.22STLYY297 pKa = 9.05WHH299 pKa = 7.11EE300 pKa = 4.13KK301 pKa = 9.32
Molecular weight: 34.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
618 |
301 |
317 |
309.0 |
35.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.987 ± 0.974 | 1.456 ± 0.335 |
7.929 ± 0.452 | 4.531 ± 0.639 |
5.178 ± 0.142 | 8.738 ± 0.557 |
2.751 ± 0.31 | 4.045 ± 0.043 |
5.178 ± 0.585 | 6.796 ± 1.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.653 | 5.016 ± 0.024 |
3.722 ± 0.776 | 2.589 ± 0.292 |
8.738 ± 1.623 | 7.12 ± 1.107 |
5.987 ± 1.933 | 4.854 ± 0.094 |
2.265 ± 0.044 | 4.693 ± 0.999 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
