
Weeksella virosa (strain ATCC 43766 / DSM 16922 / JCM 21250 / NBRC 16016 / NCTC 11634 / CL345/78)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Weeksellaceae; Weeksella; Weeksella virosa
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
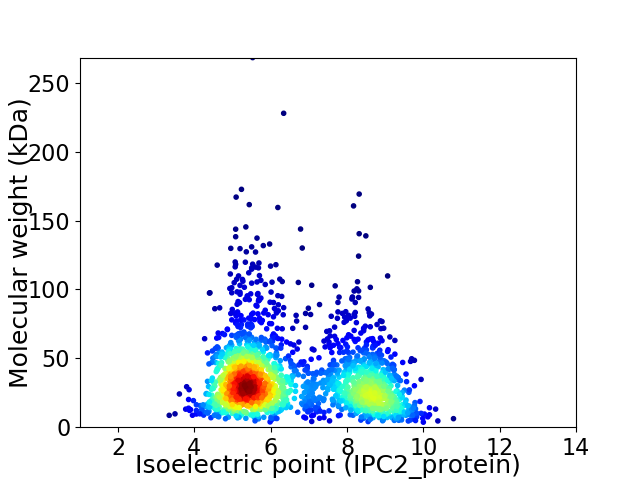
Virtual 2D-PAGE plot for 2045 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F0P2G6|F0P2G6_WEEVC Uncharacterized protein OS=Weeksella virosa (strain ATCC 43766 / DSM 16922 / JCM 21250 / NBRC 16016 / NCTC 11634 / CL345/78) OX=865938 GN=Weevi_0050 PE=4 SV=1
MM1 pKa = 7.37EE2 pKa = 5.38NKK4 pKa = 9.88TNFCEE9 pKa = 4.17EE10 pKa = 5.15DD11 pKa = 3.08ISEE14 pKa = 4.67LKK16 pKa = 10.83DD17 pKa = 3.27GLTPFPEE24 pKa = 4.21EE25 pKa = 3.86VFVAEE30 pKa = 4.23QSWLNDD36 pKa = 3.22CFLPLISVDD45 pKa = 4.0LGILRR50 pKa = 11.84TDD52 pKa = 3.73LAGTVVHH59 pKa = 6.66FLNPVEE65 pKa = 4.49PADD68 pKa = 3.84GLLGEE73 pKa = 4.45EE74 pKa = 4.16TEE76 pKa = 4.18EE77 pKa = 4.07FHH79 pKa = 8.1NEE81 pKa = 3.24FCAEE85 pKa = 3.38NWIAFKK91 pKa = 10.08LTTDD95 pKa = 2.97NKK97 pKa = 10.83YY98 pKa = 11.26NFLADD103 pKa = 3.36KK104 pKa = 10.68DD105 pKa = 4.3YY106 pKa = 11.07FLSLSEE112 pKa = 4.37CDD114 pKa = 3.24EE115 pKa = 4.77DD116 pKa = 4.51LAEE119 pKa = 5.49HH120 pKa = 6.27IQTMRR125 pKa = 11.84DD126 pKa = 2.88TFQTVKK132 pKa = 10.47SKK134 pKa = 11.17YY135 pKa = 9.22KK136 pKa = 10.68EE137 pKa = 3.92KK138 pKa = 10.77GQLLSWQDD146 pKa = 3.4YY147 pKa = 9.96PDD149 pKa = 3.78ALNFIDD155 pKa = 5.38RR156 pKa = 11.84LDD158 pKa = 3.92GEE160 pKa = 4.47ILGGNWVDD168 pKa = 3.75TVDD171 pKa = 3.4IPSAFEE177 pKa = 4.13MNFEE181 pKa = 4.36TPPEE185 pKa = 4.48DD186 pKa = 3.71SDD188 pKa = 4.26SDD190 pKa = 4.37GISISYY196 pKa = 8.91QGKK199 pKa = 7.86EE200 pKa = 3.62LMYY203 pKa = 9.31VGEE206 pKa = 4.27VAGYY210 pKa = 9.01NYY212 pKa = 10.32CSEE215 pKa = 4.21GADD218 pKa = 3.26AIMIFYY224 pKa = 10.14EE225 pKa = 3.99PEE227 pKa = 3.44NRR229 pKa = 11.84IVLFTYY235 pKa = 10.08DD236 pKa = 2.92WSS238 pKa = 3.73
MM1 pKa = 7.37EE2 pKa = 5.38NKK4 pKa = 9.88TNFCEE9 pKa = 4.17EE10 pKa = 5.15DD11 pKa = 3.08ISEE14 pKa = 4.67LKK16 pKa = 10.83DD17 pKa = 3.27GLTPFPEE24 pKa = 4.21EE25 pKa = 3.86VFVAEE30 pKa = 4.23QSWLNDD36 pKa = 3.22CFLPLISVDD45 pKa = 4.0LGILRR50 pKa = 11.84TDD52 pKa = 3.73LAGTVVHH59 pKa = 6.66FLNPVEE65 pKa = 4.49PADD68 pKa = 3.84GLLGEE73 pKa = 4.45EE74 pKa = 4.16TEE76 pKa = 4.18EE77 pKa = 4.07FHH79 pKa = 8.1NEE81 pKa = 3.24FCAEE85 pKa = 3.38NWIAFKK91 pKa = 10.08LTTDD95 pKa = 2.97NKK97 pKa = 10.83YY98 pKa = 11.26NFLADD103 pKa = 3.36KK104 pKa = 10.68DD105 pKa = 4.3YY106 pKa = 11.07FLSLSEE112 pKa = 4.37CDD114 pKa = 3.24EE115 pKa = 4.77DD116 pKa = 4.51LAEE119 pKa = 5.49HH120 pKa = 6.27IQTMRR125 pKa = 11.84DD126 pKa = 2.88TFQTVKK132 pKa = 10.47SKK134 pKa = 11.17YY135 pKa = 9.22KK136 pKa = 10.68EE137 pKa = 3.92KK138 pKa = 10.77GQLLSWQDD146 pKa = 3.4YY147 pKa = 9.96PDD149 pKa = 3.78ALNFIDD155 pKa = 5.38RR156 pKa = 11.84LDD158 pKa = 3.92GEE160 pKa = 4.47ILGGNWVDD168 pKa = 3.75TVDD171 pKa = 3.4IPSAFEE177 pKa = 4.13MNFEE181 pKa = 4.36TPPEE185 pKa = 4.48DD186 pKa = 3.71SDD188 pKa = 4.26SDD190 pKa = 4.37GISISYY196 pKa = 8.91QGKK199 pKa = 7.86EE200 pKa = 3.62LMYY203 pKa = 9.31VGEE206 pKa = 4.27VAGYY210 pKa = 9.01NYY212 pKa = 10.32CSEE215 pKa = 4.21GADD218 pKa = 3.26AIMIFYY224 pKa = 10.14EE225 pKa = 3.99PEE227 pKa = 3.44NRR229 pKa = 11.84IVLFTYY235 pKa = 10.08DD236 pKa = 2.92WSS238 pKa = 3.73
Molecular weight: 27.23 kDa
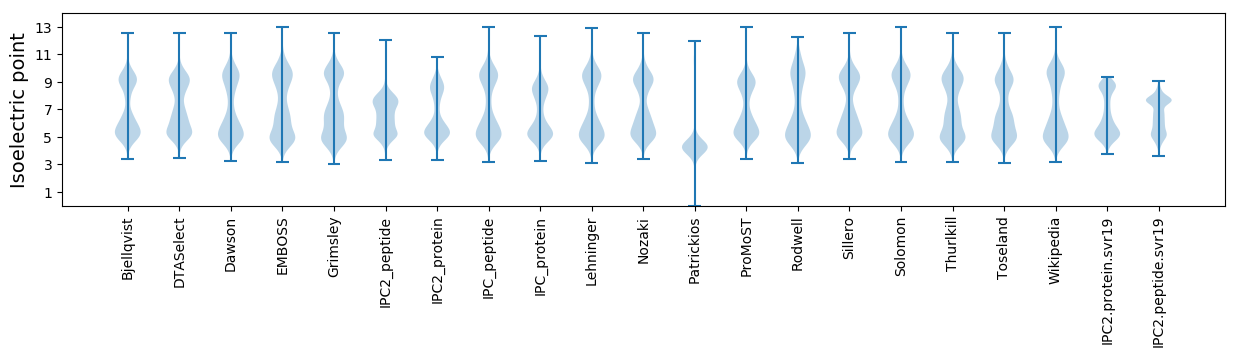
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0NXY9|F0NXY9_WEEVC Putative mannosyltransferase OS=Weeksella virosa (strain ATCC 43766 / DSM 16922 / JCM 21250 / NBRC 16016 / NCTC 11634 / CL345/78) OX=865938 GN=Weevi_1354 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.89HH16 pKa = 3.92GFRR19 pKa = 11.84EE20 pKa = 4.1RR21 pKa = 11.84MSTKK25 pKa = 9.7NGRR28 pKa = 11.84RR29 pKa = 11.84VLANRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.85KK37 pKa = 10.15GRR39 pKa = 11.84KK40 pKa = 8.68ALTISEE46 pKa = 4.15VRR48 pKa = 11.84AKK50 pKa = 10.5RR51 pKa = 3.41
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.89HH16 pKa = 3.92GFRR19 pKa = 11.84EE20 pKa = 4.1RR21 pKa = 11.84MSTKK25 pKa = 9.7NGRR28 pKa = 11.84RR29 pKa = 11.84VLANRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.85KK37 pKa = 10.15GRR39 pKa = 11.84KK40 pKa = 8.68ALTISEE46 pKa = 4.15VRR48 pKa = 11.84AKK50 pKa = 10.5RR51 pKa = 3.41
Molecular weight: 6.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
668002 |
31 |
2351 |
326.7 |
37.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.245 ± 0.05 | 0.695 ± 0.015 |
5.397 ± 0.041 | 6.908 ± 0.056 |
5.221 ± 0.044 | 6.088 ± 0.051 |
1.895 ± 0.023 | 8.056 ± 0.053 |
7.764 ± 0.05 | 9.394 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.284 ± 0.022 | 6.001 ± 0.054 |
3.411 ± 0.029 | 4.034 ± 0.035 |
3.731 ± 0.035 | 5.977 ± 0.04 |
5.514 ± 0.034 | 6.151 ± 0.047 |
1.012 ± 0.018 | 4.223 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
