
Chryseobacterium sp. FH1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Weeksellaceae; Chryseobacterium group; Chryseobacterium; unclassified Chryseobacterium
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
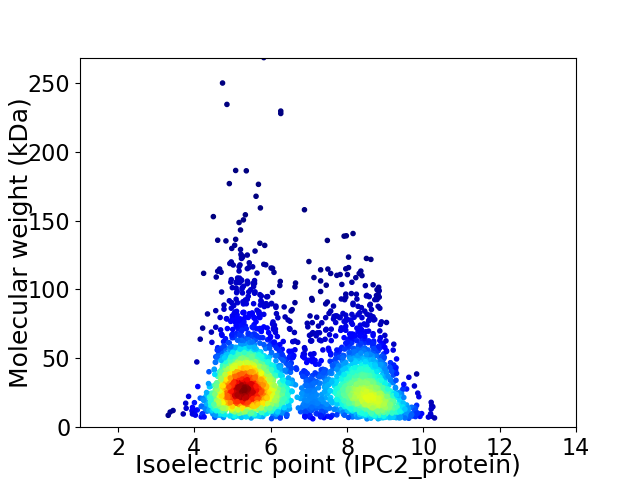
Virtual 2D-PAGE plot for 3367 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
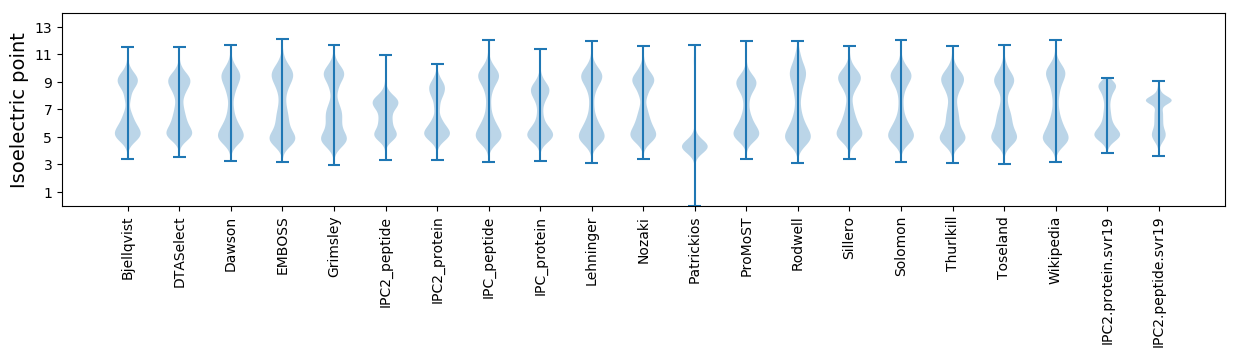
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A085B732|A0A085B732_9FLAO Amino acid racemase OS=Chryseobacterium sp. FH1 OX=1233951 GN=IO90_18840 PE=4 SV=1
MM1 pKa = 6.86KK2 pKa = 9.75TIIIGAIALLTMNFSFAQSNVDD24 pKa = 3.98LSAQIGNLNTATVDD38 pKa = 3.06QTGFMNFNALLQDD51 pKa = 3.98GNRR54 pKa = 11.84NDD56 pKa = 4.49ADD58 pKa = 3.49IDD60 pKa = 3.75QVGWGNSNLALSQGNRR76 pKa = 11.84NSIDD80 pKa = 3.13VDD82 pKa = 2.98QWGIGNSNTTSQYY95 pKa = 10.76GNRR98 pKa = 11.84NSSQTLQVGLFNDD111 pKa = 3.44VDD113 pKa = 3.71QVQIGRR119 pKa = 11.84RR120 pKa = 11.84NDD122 pKa = 3.04ASATQFGMGNTIGQYY137 pKa = 9.68QDD139 pKa = 2.72GRR141 pKa = 11.84RR142 pKa = 11.84NSATAIQVGVDD153 pKa = 3.11NTIWQDD159 pKa = 3.12QYY161 pKa = 11.49GRR163 pKa = 11.84RR164 pKa = 11.84NVAYY168 pKa = 9.7AFQAGSDD175 pKa = 3.78NYY177 pKa = 10.13IHH179 pKa = 6.62QLQDD183 pKa = 3.34GNDD186 pKa = 3.34NSATHH191 pKa = 6.21LQFGDD196 pKa = 3.37SNYY199 pKa = 10.58ADD201 pKa = 3.24SHH203 pKa = 6.82QYY205 pKa = 11.44GNDD208 pKa = 3.0NTTAGLQVGNGNEE221 pKa = 4.16LYY223 pKa = 10.31QYY225 pKa = 11.04QYY227 pKa = 11.84GNGNTAMDD235 pKa = 3.68IQMGDD240 pKa = 2.99SNYY243 pKa = 10.2TDD245 pKa = 3.37VTQTGTSHH253 pKa = 6.9LHH255 pKa = 5.87MGMQAGNNNSLVVNQSNN272 pKa = 2.98
MM1 pKa = 6.86KK2 pKa = 9.75TIIIGAIALLTMNFSFAQSNVDD24 pKa = 3.98LSAQIGNLNTATVDD38 pKa = 3.06QTGFMNFNALLQDD51 pKa = 3.98GNRR54 pKa = 11.84NDD56 pKa = 4.49ADD58 pKa = 3.49IDD60 pKa = 3.75QVGWGNSNLALSQGNRR76 pKa = 11.84NSIDD80 pKa = 3.13VDD82 pKa = 2.98QWGIGNSNTTSQYY95 pKa = 10.76GNRR98 pKa = 11.84NSSQTLQVGLFNDD111 pKa = 3.44VDD113 pKa = 3.71QVQIGRR119 pKa = 11.84RR120 pKa = 11.84NDD122 pKa = 3.04ASATQFGMGNTIGQYY137 pKa = 9.68QDD139 pKa = 2.72GRR141 pKa = 11.84RR142 pKa = 11.84NSATAIQVGVDD153 pKa = 3.11NTIWQDD159 pKa = 3.12QYY161 pKa = 11.49GRR163 pKa = 11.84RR164 pKa = 11.84NVAYY168 pKa = 9.7AFQAGSDD175 pKa = 3.78NYY177 pKa = 10.13IHH179 pKa = 6.62QLQDD183 pKa = 3.34GNDD186 pKa = 3.34NSATHH191 pKa = 6.21LQFGDD196 pKa = 3.37SNYY199 pKa = 10.58ADD201 pKa = 3.24SHH203 pKa = 6.82QYY205 pKa = 11.44GNDD208 pKa = 3.0NTTAGLQVGNGNEE221 pKa = 4.16LYY223 pKa = 10.31QYY225 pKa = 11.04QYY227 pKa = 11.84GNGNTAMDD235 pKa = 3.68IQMGDD240 pKa = 2.99SNYY243 pKa = 10.2TDD245 pKa = 3.37VTQTGTSHH253 pKa = 6.9LHH255 pKa = 5.87MGMQAGNNNSLVVNQSNN272 pKa = 2.98
Molecular weight: 29.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A085BA57|A0A085BA57_9FLAO Uncharacterized protein OS=Chryseobacterium sp. FH1 OX=1233951 GN=IO90_08585 PE=4 SV=1
MM1 pKa = 7.21TPGRR5 pKa = 11.84EE6 pKa = 4.16KK7 pKa = 10.59PPHH10 pKa = 6.29LLALAPLQRR19 pKa = 11.84RR20 pKa = 11.84GMGFFCIGYY29 pKa = 9.41ILQKK33 pKa = 8.73PSKK36 pKa = 8.46YY37 pKa = 10.71AEE39 pKa = 4.43DD40 pKa = 3.98RR41 pKa = 11.84STASGVANRR50 pKa = 11.84YY51 pKa = 7.75IRR53 pKa = 11.84KK54 pKa = 9.15ILPPTLKK61 pKa = 10.76GGIFPVGLKK70 pKa = 10.18LL71 pKa = 4.29
MM1 pKa = 7.21TPGRR5 pKa = 11.84EE6 pKa = 4.16KK7 pKa = 10.59PPHH10 pKa = 6.29LLALAPLQRR19 pKa = 11.84RR20 pKa = 11.84GMGFFCIGYY29 pKa = 9.41ILQKK33 pKa = 8.73PSKK36 pKa = 8.46YY37 pKa = 10.71AEE39 pKa = 4.43DD40 pKa = 3.98RR41 pKa = 11.84STASGVANRR50 pKa = 11.84YY51 pKa = 7.75IRR53 pKa = 11.84KK54 pKa = 9.15ILPPTLKK61 pKa = 10.76GGIFPVGLKK70 pKa = 10.18LL71 pKa = 4.29
Molecular weight: 7.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1084421 |
54 |
2373 |
322.1 |
36.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.944 ± 0.04 | 0.677 ± 0.011 |
5.575 ± 0.031 | 6.78 ± 0.049 |
5.584 ± 0.036 | 6.176 ± 0.046 |
1.58 ± 0.02 | 8.053 ± 0.043 |
8.443 ± 0.046 | 9.136 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.217 ± 0.023 | 6.538 ± 0.045 |
3.3 ± 0.022 | 3.502 ± 0.024 |
3.226 ± 0.023 | 6.755 ± 0.036 |
5.439 ± 0.041 | 5.946 ± 0.031 |
1.026 ± 0.014 | 4.103 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
