
Propionispora sp. 2/2-37
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Negativicutes; Selenomonadales; Sporomusaceae; Propionispora; unclassified Propionispora
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
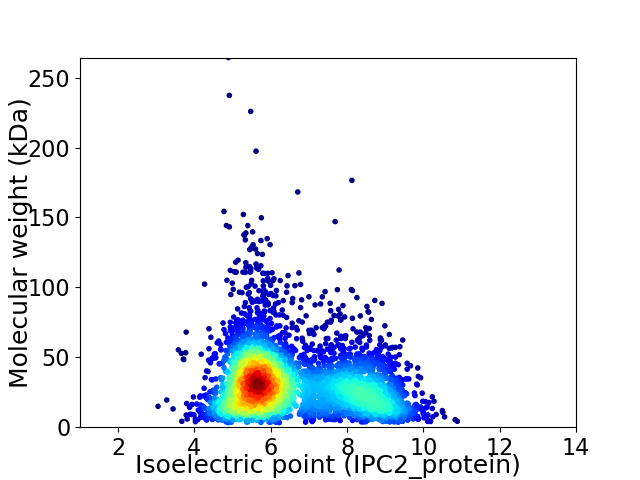
Virtual 2D-PAGE plot for 3949 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
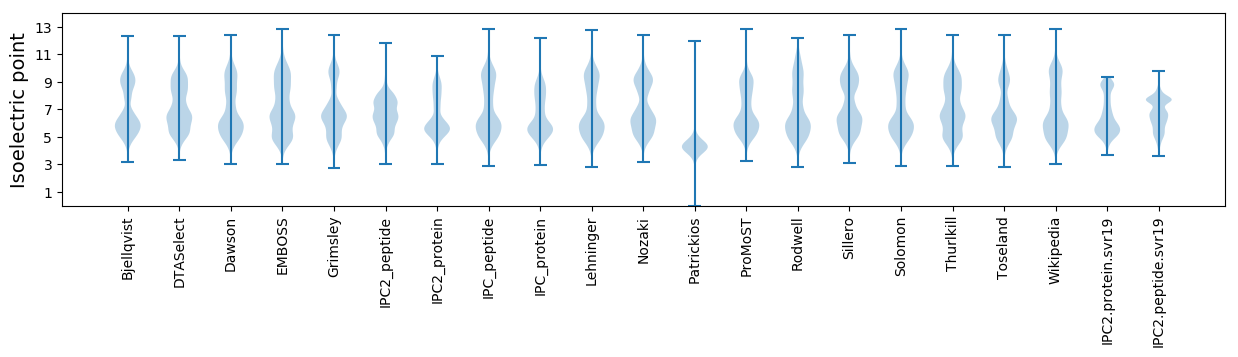
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K8JFJ5|A0A0K8JFJ5_9FIRM Protein-glutamate methylesterase/protein-glutamine glutaminase OS=Propionispora sp. 2/2-37 OX=1677858 GN=cheB PE=3 SV=1
MM1 pKa = 7.55LRR3 pKa = 11.84ITDD6 pKa = 3.84SMVTAQTLANINNNTKK22 pKa = 10.32RR23 pKa = 11.84LSEE26 pKa = 4.22ANDD29 pKa = 3.15RR30 pKa = 11.84VASGKK35 pKa = 10.69KK36 pKa = 9.03IDD38 pKa = 4.42EE39 pKa = 4.8ASDD42 pKa = 3.86DD43 pKa = 3.76PTVAARR49 pKa = 11.84AIKK52 pKa = 9.14YY53 pKa = 9.32RR54 pKa = 11.84NYY56 pKa = 9.14VAKK59 pKa = 10.4IEE61 pKa = 4.05QYY63 pKa = 10.76QDD65 pKa = 2.87NASAATSWQKK75 pKa = 9.38VTDD78 pKa = 4.12DD79 pKa = 5.5AMDD82 pKa = 4.79DD83 pKa = 3.92LDD85 pKa = 6.15DD86 pKa = 4.64VIQRR90 pKa = 11.84LQEE93 pKa = 3.78ITTQAASTATSSDD106 pKa = 2.95TDD108 pKa = 3.64MEE110 pKa = 4.36ALQQEE115 pKa = 4.75VTEE118 pKa = 4.45LQQQLVDD125 pKa = 3.94ILNTSYY131 pKa = 11.2AGRR134 pKa = 11.84YY135 pKa = 7.66VFAGYY140 pKa = 7.95DD141 pKa = 3.31TGEE144 pKa = 3.9APYY147 pKa = 11.2ALVQTYY153 pKa = 7.65TAADD157 pKa = 3.59VTTTDD162 pKa = 3.37DD163 pKa = 2.9TGYY166 pKa = 9.2TEE168 pKa = 5.45DD169 pKa = 4.36SIGGITGLTAGSSYY183 pKa = 10.56TLTVADD189 pKa = 3.65STAYY193 pKa = 10.27AGAYY197 pKa = 10.13SYY199 pKa = 10.44TLTDD203 pKa = 3.35ANGNIVASVDD213 pKa = 3.73NITDD217 pKa = 3.28TTTAVTLTGTDD228 pKa = 3.81DD229 pKa = 4.02EE230 pKa = 5.72SITLAGTTGVTTGDD244 pKa = 3.08MTFTVNNSSLGSTITYY260 pKa = 9.34KK261 pKa = 11.06GQVLSLCGVVDD272 pKa = 4.31SSVSDD277 pKa = 4.16DD278 pKa = 5.53DD279 pKa = 4.42IISYY283 pKa = 7.97YY284 pKa = 8.73TANASVEE291 pKa = 3.91YY292 pKa = 10.49DD293 pKa = 3.24AANSGTQAIKK303 pKa = 10.92YY304 pKa = 8.55NIGYY308 pKa = 9.79GIEE311 pKa = 3.72IAVNVEE317 pKa = 3.96GQDD320 pKa = 3.39ITGTGADD327 pKa = 4.15DD328 pKa = 3.65TNLFDD333 pKa = 4.46TVAKK337 pKa = 10.59LLLSLDD343 pKa = 3.59GDD345 pKa = 4.18TSYY348 pKa = 9.41KK349 pKa = 9.84TYY351 pKa = 10.82NSEE354 pKa = 3.88TGEE357 pKa = 4.18VEE359 pKa = 4.44TVSFTTDD366 pKa = 2.82EE367 pKa = 4.85LLTEE371 pKa = 4.43LQTNEE376 pKa = 4.25DD377 pKa = 3.63RR378 pKa = 11.84LLTTRR383 pKa = 11.84TDD385 pKa = 3.13LGARR389 pKa = 11.84MSYY392 pKa = 9.97TSTVTSRR399 pKa = 11.84LSNDD403 pKa = 2.5LTTYY407 pKa = 11.02KK408 pKa = 10.37EE409 pKa = 4.22FKK411 pKa = 10.35SDD413 pKa = 4.23NEE415 pKa = 4.2DD416 pKa = 2.91VDD418 pKa = 3.45IAEE421 pKa = 4.82AATEE425 pKa = 4.06QATAEE430 pKa = 4.16YY431 pKa = 10.2VYY433 pKa = 10.44QASLTVGASVISKK446 pKa = 8.65TLVDD450 pKa = 3.86YY451 pKa = 10.65LAA453 pKa = 5.2
MM1 pKa = 7.55LRR3 pKa = 11.84ITDD6 pKa = 3.84SMVTAQTLANINNNTKK22 pKa = 10.32RR23 pKa = 11.84LSEE26 pKa = 4.22ANDD29 pKa = 3.15RR30 pKa = 11.84VASGKK35 pKa = 10.69KK36 pKa = 9.03IDD38 pKa = 4.42EE39 pKa = 4.8ASDD42 pKa = 3.86DD43 pKa = 3.76PTVAARR49 pKa = 11.84AIKK52 pKa = 9.14YY53 pKa = 9.32RR54 pKa = 11.84NYY56 pKa = 9.14VAKK59 pKa = 10.4IEE61 pKa = 4.05QYY63 pKa = 10.76QDD65 pKa = 2.87NASAATSWQKK75 pKa = 9.38VTDD78 pKa = 4.12DD79 pKa = 5.5AMDD82 pKa = 4.79DD83 pKa = 3.92LDD85 pKa = 6.15DD86 pKa = 4.64VIQRR90 pKa = 11.84LQEE93 pKa = 3.78ITTQAASTATSSDD106 pKa = 2.95TDD108 pKa = 3.64MEE110 pKa = 4.36ALQQEE115 pKa = 4.75VTEE118 pKa = 4.45LQQQLVDD125 pKa = 3.94ILNTSYY131 pKa = 11.2AGRR134 pKa = 11.84YY135 pKa = 7.66VFAGYY140 pKa = 7.95DD141 pKa = 3.31TGEE144 pKa = 3.9APYY147 pKa = 11.2ALVQTYY153 pKa = 7.65TAADD157 pKa = 3.59VTTTDD162 pKa = 3.37DD163 pKa = 2.9TGYY166 pKa = 9.2TEE168 pKa = 5.45DD169 pKa = 4.36SIGGITGLTAGSSYY183 pKa = 10.56TLTVADD189 pKa = 3.65STAYY193 pKa = 10.27AGAYY197 pKa = 10.13SYY199 pKa = 10.44TLTDD203 pKa = 3.35ANGNIVASVDD213 pKa = 3.73NITDD217 pKa = 3.28TTTAVTLTGTDD228 pKa = 3.81DD229 pKa = 4.02EE230 pKa = 5.72SITLAGTTGVTTGDD244 pKa = 3.08MTFTVNNSSLGSTITYY260 pKa = 9.34KK261 pKa = 11.06GQVLSLCGVVDD272 pKa = 4.31SSVSDD277 pKa = 4.16DD278 pKa = 5.53DD279 pKa = 4.42IISYY283 pKa = 7.97YY284 pKa = 8.73TANASVEE291 pKa = 3.91YY292 pKa = 10.49DD293 pKa = 3.24AANSGTQAIKK303 pKa = 10.92YY304 pKa = 8.55NIGYY308 pKa = 9.79GIEE311 pKa = 3.72IAVNVEE317 pKa = 3.96GQDD320 pKa = 3.39ITGTGADD327 pKa = 4.15DD328 pKa = 3.65TNLFDD333 pKa = 4.46TVAKK337 pKa = 10.59LLLSLDD343 pKa = 3.59GDD345 pKa = 4.18TSYY348 pKa = 9.41KK349 pKa = 9.84TYY351 pKa = 10.82NSEE354 pKa = 3.88TGEE357 pKa = 4.18VEE359 pKa = 4.44TVSFTTDD366 pKa = 2.82EE367 pKa = 4.85LLTEE371 pKa = 4.43LQTNEE376 pKa = 4.25DD377 pKa = 3.63RR378 pKa = 11.84LLTTRR383 pKa = 11.84TDD385 pKa = 3.13LGARR389 pKa = 11.84MSYY392 pKa = 9.97TSTVTSRR399 pKa = 11.84LSNDD403 pKa = 2.5LTTYY407 pKa = 11.02KK408 pKa = 10.37EE409 pKa = 4.22FKK411 pKa = 10.35SDD413 pKa = 4.23NEE415 pKa = 4.2DD416 pKa = 2.91VDD418 pKa = 3.45IAEE421 pKa = 4.82AATEE425 pKa = 4.06QATAEE430 pKa = 4.16YY431 pKa = 10.2VYY433 pKa = 10.44QASLTVGASVISKK446 pKa = 8.65TLVDD450 pKa = 3.86YY451 pKa = 10.65LAA453 pKa = 5.2
Molecular weight: 48.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K8JAE7|A0A0K8JAE7_9FIRM Glucitol/sorbitol permease IIC component OS=Propionispora sp. 2/2-37 OX=1677858 GN=srlA PE=4 SV=1
MM1 pKa = 7.85WEE3 pKa = 4.25MLLLAAGLWGLQGVLGFFQMRR24 pKa = 11.84NFNGRR29 pKa = 11.84FKK31 pKa = 11.12ALRR34 pKa = 11.84QRR36 pKa = 11.84GRR38 pKa = 11.84VVAGRR43 pKa = 11.84SKK45 pKa = 9.99GRR47 pKa = 11.84MVAGVVVLFCIDD59 pKa = 3.32ANGTILTGEE68 pKa = 4.15RR69 pKa = 11.84MSGVSSFARR78 pKa = 11.84FHH80 pKa = 6.8PFPALQGNQLNKK92 pKa = 9.43IDD94 pKa = 4.1EE95 pKa = 4.86TICEE99 pKa = 4.44YY100 pKa = 10.61IRR102 pKa = 11.84LDD104 pKa = 3.57RR105 pKa = 11.84QTTRR109 pKa = 11.84AVLNAVDD116 pKa = 4.01NYY118 pKa = 11.06RR119 pKa = 11.84AFIQANTTMEE129 pKa = 4.48VIPAKK134 pKa = 10.06TGALLPP140 pKa = 4.51
MM1 pKa = 7.85WEE3 pKa = 4.25MLLLAAGLWGLQGVLGFFQMRR24 pKa = 11.84NFNGRR29 pKa = 11.84FKK31 pKa = 11.12ALRR34 pKa = 11.84QRR36 pKa = 11.84GRR38 pKa = 11.84VVAGRR43 pKa = 11.84SKK45 pKa = 9.99GRR47 pKa = 11.84MVAGVVVLFCIDD59 pKa = 3.32ANGTILTGEE68 pKa = 4.15RR69 pKa = 11.84MSGVSSFARR78 pKa = 11.84FHH80 pKa = 6.8PFPALQGNQLNKK92 pKa = 9.43IDD94 pKa = 4.1EE95 pKa = 4.86TICEE99 pKa = 4.44YY100 pKa = 10.61IRR102 pKa = 11.84LDD104 pKa = 3.57RR105 pKa = 11.84QTTRR109 pKa = 11.84AVLNAVDD116 pKa = 4.01NYY118 pKa = 11.06RR119 pKa = 11.84AFIQANTTMEE129 pKa = 4.48VIPAKK134 pKa = 10.06TGALLPP140 pKa = 4.51
Molecular weight: 15.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1189072 |
29 |
2502 |
301.1 |
33.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.888 ± 0.045 | 1.172 ± 0.016 |
4.904 ± 0.029 | 6.408 ± 0.037 |
3.931 ± 0.028 | 7.491 ± 0.044 |
1.952 ± 0.018 | 7.394 ± 0.035 |
5.86 ± 0.032 | 9.877 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.77 ± 0.02 | 4.026 ± 0.03 |
3.83 ± 0.026 | 3.916 ± 0.029 |
4.829 ± 0.031 | 5.615 ± 0.032 |
5.342 ± 0.033 | 7.441 ± 0.035 |
0.975 ± 0.014 | 3.379 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
