
Beihai weivirus-like virus 8
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.62
Get precalculated fractions of proteins
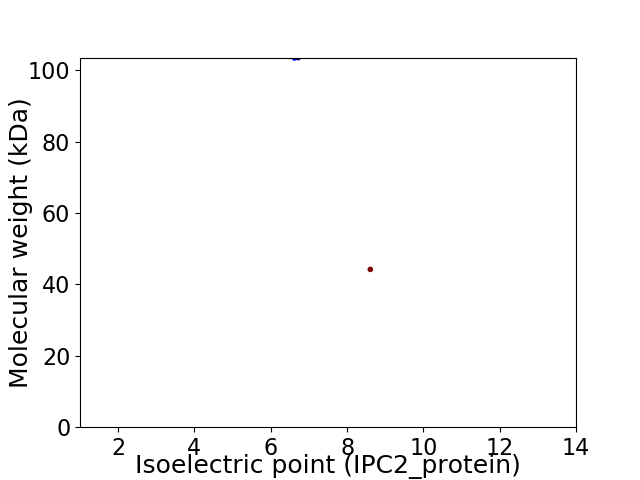
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL95|A0A1L3KL95_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 8 OX=1922755 PE=4 SV=1
MM1 pKa = 7.44TLQLSRR7 pKa = 11.84ANTTGIRR14 pKa = 11.84DD15 pKa = 3.48VEE17 pKa = 3.79PWMFKK22 pKa = 9.95IARR25 pKa = 11.84EE26 pKa = 3.93RR27 pKa = 11.84PGMYY31 pKa = 9.09TIKK34 pKa = 10.36IFIPSMPDD42 pKa = 3.07CPFDD46 pKa = 3.28IAKK49 pKa = 10.41AEE51 pKa = 4.14SVMRR55 pKa = 11.84KK56 pKa = 8.48VLLYY60 pKa = 10.93GSVHH64 pKa = 7.12LLLCPVAHH72 pKa = 7.04LMLGMPGFFYY82 pKa = 10.8FLAVVNVIGFVFVYY96 pKa = 7.11TQWPEE101 pKa = 4.24TIGEE105 pKa = 4.22TLEE108 pKa = 4.09VNFTGEE114 pKa = 4.03EE115 pKa = 3.89FAKK118 pKa = 10.23LDD120 pKa = 3.86KK121 pKa = 11.24VIMNQTYY128 pKa = 8.46TEE130 pKa = 4.37SKK132 pKa = 10.67SKK134 pKa = 9.94VQSAIVFQHH143 pKa = 6.72ASVIWPSHH151 pKa = 5.47TPDD154 pKa = 3.84DD155 pKa = 3.72VTMRR159 pKa = 11.84TLGPWAITEE168 pKa = 4.07YY169 pKa = 10.27RR170 pKa = 11.84CNGTWMKK177 pKa = 11.05NFFKK181 pKa = 10.67RR182 pKa = 11.84FPYY185 pKa = 10.16LPCGYY190 pKa = 10.64SMKK193 pKa = 10.51KK194 pKa = 7.81QVRR197 pKa = 11.84LCLRR201 pKa = 11.84QGIGGATVTSKK212 pKa = 11.25DD213 pKa = 3.08GDD215 pKa = 3.94LPEE218 pKa = 5.73ADD220 pKa = 4.54PTPKK224 pKa = 9.97PAPPVDD230 pKa = 3.97LSSDD234 pKa = 3.32TGVPVDD240 pKa = 3.38KK241 pKa = 10.56RR242 pKa = 11.84YY243 pKa = 10.68VRR245 pKa = 11.84TQDD248 pKa = 2.74VSQIEE253 pKa = 4.25RR254 pKa = 11.84HH255 pKa = 4.51AHH257 pKa = 5.17KK258 pKa = 10.49CCEE261 pKa = 4.13CGCTYY266 pKa = 10.99EE267 pKa = 4.56HH268 pKa = 6.73GHH270 pKa = 6.7PGGHH274 pKa = 6.29GKK276 pKa = 9.61HH277 pKa = 5.53GQCLGNCPEE286 pKa = 4.3PTCPRR291 pKa = 11.84HH292 pKa = 6.96DD293 pKa = 4.37PGVDD297 pKa = 3.17YY298 pKa = 11.4AGAQKK303 pKa = 10.42LQPTPPKK310 pKa = 9.96EE311 pKa = 4.15EE312 pKa = 4.3PKK314 pKa = 8.4TTEE317 pKa = 3.91SKK319 pKa = 10.68PPRR322 pKa = 11.84NDD324 pKa = 2.92LGYY327 pKa = 10.48VEE329 pKa = 5.4PGHH332 pKa = 6.56GNEE335 pKa = 4.92DD336 pKa = 3.65LHH338 pKa = 7.35PLGGASAEE346 pKa = 4.14DD347 pKa = 3.67TEE349 pKa = 5.02IRR351 pKa = 11.84AEE353 pKa = 3.96RR354 pKa = 11.84QDD356 pKa = 4.01RR357 pKa = 11.84IVEE360 pKa = 4.1NVAGVGVVGQSTDD373 pKa = 2.83SSNTKK378 pKa = 10.19QIVGVVSLPVTDD390 pKa = 4.0TPNVYY395 pKa = 10.42ARR397 pKa = 11.84EE398 pKa = 4.04AANVQSAIDD407 pKa = 3.49NRR409 pKa = 11.84ITKK412 pKa = 9.11KK413 pKa = 10.28QRR415 pKa = 11.84PFTANKK421 pKa = 9.09EE422 pKa = 4.14DD423 pKa = 3.62KK424 pKa = 10.72ALLGRR429 pKa = 11.84LVYY432 pKa = 10.32AAIGDD437 pKa = 3.8HH438 pKa = 6.34PRR440 pKa = 11.84RR441 pKa = 11.84SLFSARR447 pKa = 11.84RR448 pKa = 11.84VTAWWEE454 pKa = 3.78THH456 pKa = 6.39LFADD460 pKa = 4.81LKK462 pKa = 9.91SGKK465 pKa = 6.74WTEE468 pKa = 4.25DD469 pKa = 2.98RR470 pKa = 11.84LSRR473 pKa = 11.84AIEE476 pKa = 4.0NLCGRR481 pKa = 11.84IQPGFKK487 pKa = 9.98LACDD491 pKa = 3.82VKK493 pKa = 10.74LEE495 pKa = 4.01PMPEE499 pKa = 3.92GKK501 pKa = 10.26APRR504 pKa = 11.84MLIADD509 pKa = 3.75GDD511 pKa = 4.09EE512 pKa = 4.48GQVLALLTVCCIEE525 pKa = 5.12DD526 pKa = 3.89SIKK529 pKa = 10.83KK530 pKa = 9.47HH531 pKa = 5.8LPKK534 pKa = 9.8KK535 pKa = 8.18TIKK538 pKa = 10.87GLGKK542 pKa = 8.82RR543 pKa = 11.84AAMEE547 pKa = 3.73RR548 pKa = 11.84VAAEE552 pKa = 3.7LRR554 pKa = 11.84APKK557 pKa = 9.94AAYY560 pKa = 10.58SNTKK564 pKa = 7.57TQSTQRR570 pKa = 11.84TGNGKK575 pKa = 7.52MLKK578 pKa = 9.3PGVSIFEE585 pKa = 4.53GDD587 pKa = 3.13GSAWDD592 pKa = 4.16TTCSIQLRR600 pKa = 11.84DD601 pKa = 3.75CVEE604 pKa = 4.01NPVILHH610 pKa = 5.91VGSILKK616 pKa = 10.24VFMAQPQSWIQAHH629 pKa = 7.04DD630 pKa = 5.0DD631 pKa = 3.31ISVLDD636 pKa = 3.94KK637 pKa = 10.86LTITFKK643 pKa = 11.2KK644 pKa = 9.7NGEE647 pKa = 4.05FRR649 pKa = 11.84KK650 pKa = 10.28HH651 pKa = 6.4IIDD654 pKa = 4.97AIRR657 pKa = 11.84RR658 pKa = 11.84SGHH661 pKa = 6.55RR662 pKa = 11.84GTSCLNWWINFTCWHH677 pKa = 6.1CAIFEE682 pKa = 4.16EE683 pKa = 5.05PEE685 pKa = 4.13KK686 pKa = 10.94FLDD689 pKa = 3.52PDD691 pKa = 3.12VRR693 pKa = 11.84YY694 pKa = 10.4GRR696 pKa = 11.84DD697 pKa = 2.88HH698 pKa = 7.03AGTYY702 pKa = 9.42RR703 pKa = 11.84WIASAFEE710 pKa = 4.57GDD712 pKa = 4.43DD713 pKa = 4.47SILSTTPRR721 pKa = 11.84IEE723 pKa = 4.44EE724 pKa = 3.81KK725 pKa = 10.91DD726 pKa = 3.52EE727 pKa = 4.95LYY729 pKa = 11.05VSLMQRR735 pKa = 11.84WEE737 pKa = 3.86RR738 pKa = 11.84FGFNMKK744 pKa = 9.92IFIRR748 pKa = 11.84AKK750 pKa = 8.55RR751 pKa = 11.84ALFTGYY757 pKa = 10.67HH758 pKa = 5.56MALDD762 pKa = 3.98NDD764 pKa = 4.25GPTGVLMPEE773 pKa = 3.99VDD775 pKa = 3.19RR776 pKa = 11.84CFARR780 pKa = 11.84AGISCSPSMIEE791 pKa = 3.86YY792 pKa = 9.88FKK794 pKa = 11.21KK795 pKa = 10.47GDD797 pKa = 3.46RR798 pKa = 11.84AGCQSVSRR806 pKa = 11.84AAALSRR812 pKa = 11.84AHH814 pKa = 6.76EE815 pKa = 4.32FAGCAPTISTKK826 pKa = 7.95YY827 pKa = 9.13LRR829 pKa = 11.84YY830 pKa = 9.77YY831 pKa = 10.52EE832 pKa = 4.14SLAVKK837 pKa = 9.74TNVDD841 pKa = 3.15RR842 pKa = 11.84DD843 pKa = 3.55LQMRR847 pKa = 11.84TCGGDD852 pKa = 3.37AEE854 pKa = 4.64FSEE857 pKa = 4.62PDD859 pKa = 2.99IVAEE863 pKa = 4.09INLKK867 pKa = 10.53NGAAMTFDD875 pKa = 3.05SAEE878 pKa = 4.14RR879 pKa = 11.84DD880 pKa = 3.12RR881 pKa = 11.84LAAVGFEE888 pKa = 4.25CTEE891 pKa = 4.09EE892 pKa = 3.99EE893 pKa = 4.32LSRR896 pKa = 11.84FSLRR900 pKa = 11.84VWDD903 pKa = 3.96YY904 pKa = 11.89DD905 pKa = 4.16VLKK908 pKa = 10.32DD909 pKa = 3.35WEE911 pKa = 4.63GFRR914 pKa = 11.84EE915 pKa = 4.15SLPEE919 pKa = 3.87SWRR922 pKa = 11.84MAA924 pKa = 3.76
MM1 pKa = 7.44TLQLSRR7 pKa = 11.84ANTTGIRR14 pKa = 11.84DD15 pKa = 3.48VEE17 pKa = 3.79PWMFKK22 pKa = 9.95IARR25 pKa = 11.84EE26 pKa = 3.93RR27 pKa = 11.84PGMYY31 pKa = 9.09TIKK34 pKa = 10.36IFIPSMPDD42 pKa = 3.07CPFDD46 pKa = 3.28IAKK49 pKa = 10.41AEE51 pKa = 4.14SVMRR55 pKa = 11.84KK56 pKa = 8.48VLLYY60 pKa = 10.93GSVHH64 pKa = 7.12LLLCPVAHH72 pKa = 7.04LMLGMPGFFYY82 pKa = 10.8FLAVVNVIGFVFVYY96 pKa = 7.11TQWPEE101 pKa = 4.24TIGEE105 pKa = 4.22TLEE108 pKa = 4.09VNFTGEE114 pKa = 4.03EE115 pKa = 3.89FAKK118 pKa = 10.23LDD120 pKa = 3.86KK121 pKa = 11.24VIMNQTYY128 pKa = 8.46TEE130 pKa = 4.37SKK132 pKa = 10.67SKK134 pKa = 9.94VQSAIVFQHH143 pKa = 6.72ASVIWPSHH151 pKa = 5.47TPDD154 pKa = 3.84DD155 pKa = 3.72VTMRR159 pKa = 11.84TLGPWAITEE168 pKa = 4.07YY169 pKa = 10.27RR170 pKa = 11.84CNGTWMKK177 pKa = 11.05NFFKK181 pKa = 10.67RR182 pKa = 11.84FPYY185 pKa = 10.16LPCGYY190 pKa = 10.64SMKK193 pKa = 10.51KK194 pKa = 7.81QVRR197 pKa = 11.84LCLRR201 pKa = 11.84QGIGGATVTSKK212 pKa = 11.25DD213 pKa = 3.08GDD215 pKa = 3.94LPEE218 pKa = 5.73ADD220 pKa = 4.54PTPKK224 pKa = 9.97PAPPVDD230 pKa = 3.97LSSDD234 pKa = 3.32TGVPVDD240 pKa = 3.38KK241 pKa = 10.56RR242 pKa = 11.84YY243 pKa = 10.68VRR245 pKa = 11.84TQDD248 pKa = 2.74VSQIEE253 pKa = 4.25RR254 pKa = 11.84HH255 pKa = 4.51AHH257 pKa = 5.17KK258 pKa = 10.49CCEE261 pKa = 4.13CGCTYY266 pKa = 10.99EE267 pKa = 4.56HH268 pKa = 6.73GHH270 pKa = 6.7PGGHH274 pKa = 6.29GKK276 pKa = 9.61HH277 pKa = 5.53GQCLGNCPEE286 pKa = 4.3PTCPRR291 pKa = 11.84HH292 pKa = 6.96DD293 pKa = 4.37PGVDD297 pKa = 3.17YY298 pKa = 11.4AGAQKK303 pKa = 10.42LQPTPPKK310 pKa = 9.96EE311 pKa = 4.15EE312 pKa = 4.3PKK314 pKa = 8.4TTEE317 pKa = 3.91SKK319 pKa = 10.68PPRR322 pKa = 11.84NDD324 pKa = 2.92LGYY327 pKa = 10.48VEE329 pKa = 5.4PGHH332 pKa = 6.56GNEE335 pKa = 4.92DD336 pKa = 3.65LHH338 pKa = 7.35PLGGASAEE346 pKa = 4.14DD347 pKa = 3.67TEE349 pKa = 5.02IRR351 pKa = 11.84AEE353 pKa = 3.96RR354 pKa = 11.84QDD356 pKa = 4.01RR357 pKa = 11.84IVEE360 pKa = 4.1NVAGVGVVGQSTDD373 pKa = 2.83SSNTKK378 pKa = 10.19QIVGVVSLPVTDD390 pKa = 4.0TPNVYY395 pKa = 10.42ARR397 pKa = 11.84EE398 pKa = 4.04AANVQSAIDD407 pKa = 3.49NRR409 pKa = 11.84ITKK412 pKa = 9.11KK413 pKa = 10.28QRR415 pKa = 11.84PFTANKK421 pKa = 9.09EE422 pKa = 4.14DD423 pKa = 3.62KK424 pKa = 10.72ALLGRR429 pKa = 11.84LVYY432 pKa = 10.32AAIGDD437 pKa = 3.8HH438 pKa = 6.34PRR440 pKa = 11.84RR441 pKa = 11.84SLFSARR447 pKa = 11.84RR448 pKa = 11.84VTAWWEE454 pKa = 3.78THH456 pKa = 6.39LFADD460 pKa = 4.81LKK462 pKa = 9.91SGKK465 pKa = 6.74WTEE468 pKa = 4.25DD469 pKa = 2.98RR470 pKa = 11.84LSRR473 pKa = 11.84AIEE476 pKa = 4.0NLCGRR481 pKa = 11.84IQPGFKK487 pKa = 9.98LACDD491 pKa = 3.82VKK493 pKa = 10.74LEE495 pKa = 4.01PMPEE499 pKa = 3.92GKK501 pKa = 10.26APRR504 pKa = 11.84MLIADD509 pKa = 3.75GDD511 pKa = 4.09EE512 pKa = 4.48GQVLALLTVCCIEE525 pKa = 5.12DD526 pKa = 3.89SIKK529 pKa = 10.83KK530 pKa = 9.47HH531 pKa = 5.8LPKK534 pKa = 9.8KK535 pKa = 8.18TIKK538 pKa = 10.87GLGKK542 pKa = 8.82RR543 pKa = 11.84AAMEE547 pKa = 3.73RR548 pKa = 11.84VAAEE552 pKa = 3.7LRR554 pKa = 11.84APKK557 pKa = 9.94AAYY560 pKa = 10.58SNTKK564 pKa = 7.57TQSTQRR570 pKa = 11.84TGNGKK575 pKa = 7.52MLKK578 pKa = 9.3PGVSIFEE585 pKa = 4.53GDD587 pKa = 3.13GSAWDD592 pKa = 4.16TTCSIQLRR600 pKa = 11.84DD601 pKa = 3.75CVEE604 pKa = 4.01NPVILHH610 pKa = 5.91VGSILKK616 pKa = 10.24VFMAQPQSWIQAHH629 pKa = 7.04DD630 pKa = 5.0DD631 pKa = 3.31ISVLDD636 pKa = 3.94KK637 pKa = 10.86LTITFKK643 pKa = 11.2KK644 pKa = 9.7NGEE647 pKa = 4.05FRR649 pKa = 11.84KK650 pKa = 10.28HH651 pKa = 6.4IIDD654 pKa = 4.97AIRR657 pKa = 11.84RR658 pKa = 11.84SGHH661 pKa = 6.55RR662 pKa = 11.84GTSCLNWWINFTCWHH677 pKa = 6.1CAIFEE682 pKa = 4.16EE683 pKa = 5.05PEE685 pKa = 4.13KK686 pKa = 10.94FLDD689 pKa = 3.52PDD691 pKa = 3.12VRR693 pKa = 11.84YY694 pKa = 10.4GRR696 pKa = 11.84DD697 pKa = 2.88HH698 pKa = 7.03AGTYY702 pKa = 9.42RR703 pKa = 11.84WIASAFEE710 pKa = 4.57GDD712 pKa = 4.43DD713 pKa = 4.47SILSTTPRR721 pKa = 11.84IEE723 pKa = 4.44EE724 pKa = 3.81KK725 pKa = 10.91DD726 pKa = 3.52EE727 pKa = 4.95LYY729 pKa = 11.05VSLMQRR735 pKa = 11.84WEE737 pKa = 3.86RR738 pKa = 11.84FGFNMKK744 pKa = 9.92IFIRR748 pKa = 11.84AKK750 pKa = 8.55RR751 pKa = 11.84ALFTGYY757 pKa = 10.67HH758 pKa = 5.56MALDD762 pKa = 3.98NDD764 pKa = 4.25GPTGVLMPEE773 pKa = 3.99VDD775 pKa = 3.19RR776 pKa = 11.84CFARR780 pKa = 11.84AGISCSPSMIEE791 pKa = 3.86YY792 pKa = 9.88FKK794 pKa = 11.21KK795 pKa = 10.47GDD797 pKa = 3.46RR798 pKa = 11.84AGCQSVSRR806 pKa = 11.84AAALSRR812 pKa = 11.84AHH814 pKa = 6.76EE815 pKa = 4.32FAGCAPTISTKK826 pKa = 7.95YY827 pKa = 9.13LRR829 pKa = 11.84YY830 pKa = 9.77YY831 pKa = 10.52EE832 pKa = 4.14SLAVKK837 pKa = 9.74TNVDD841 pKa = 3.15RR842 pKa = 11.84DD843 pKa = 3.55LQMRR847 pKa = 11.84TCGGDD852 pKa = 3.37AEE854 pKa = 4.64FSEE857 pKa = 4.62PDD859 pKa = 2.99IVAEE863 pKa = 4.09INLKK867 pKa = 10.53NGAAMTFDD875 pKa = 3.05SAEE878 pKa = 4.14RR879 pKa = 11.84DD880 pKa = 3.12RR881 pKa = 11.84LAAVGFEE888 pKa = 4.25CTEE891 pKa = 4.09EE892 pKa = 3.99EE893 pKa = 4.32LSRR896 pKa = 11.84FSLRR900 pKa = 11.84VWDD903 pKa = 3.96YY904 pKa = 11.89DD905 pKa = 4.16VLKK908 pKa = 10.32DD909 pKa = 3.35WEE911 pKa = 4.63GFRR914 pKa = 11.84EE915 pKa = 4.15SLPEE919 pKa = 3.87SWRR922 pKa = 11.84MAA924 pKa = 3.76
Molecular weight: 103.4 kDa
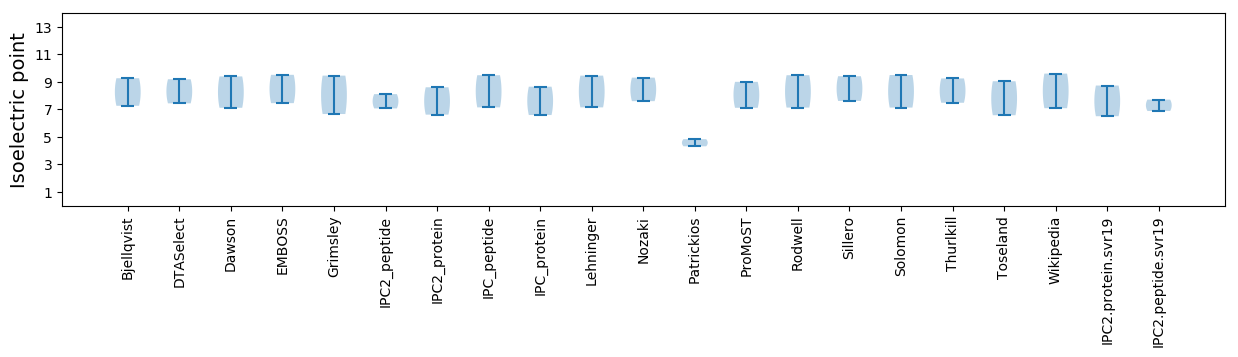
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL95|A0A1L3KL95_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 8 OX=1922755 PE=4 SV=1
MM1 pKa = 7.72ALTAKK6 pKa = 7.91QQRR9 pKa = 11.84AIADD13 pKa = 3.55AAPAAKK19 pKa = 10.06AAMRR23 pKa = 11.84AAFALQNRR31 pKa = 11.84GRR33 pKa = 11.84VPGVDD38 pKa = 3.35RR39 pKa = 11.84NAKK42 pKa = 6.97QARR45 pKa = 11.84ARR47 pKa = 11.84GRR49 pKa = 11.84RR50 pKa = 11.84GIPTAIQNMPRR61 pKa = 11.84GAAGPRR67 pKa = 11.84GEE69 pKa = 4.8MPDD72 pKa = 2.99ATRR75 pKa = 11.84FHH77 pKa = 7.45WDD79 pKa = 3.11QTPASSNVVAPRR91 pKa = 11.84GFGYY95 pKa = 10.95YY96 pKa = 10.19DD97 pKa = 4.28AFTHH101 pKa = 6.05SPSDD105 pKa = 3.23AVTAFSVGPATPIQANTRR123 pKa = 11.84ANVTTSAPDD132 pKa = 3.55GVGAGDD138 pKa = 4.82LGLSMIIVYY147 pKa = 8.74PAATDD152 pKa = 3.39AQAALFKK159 pKa = 10.84CGATATDD166 pKa = 4.4PVTTTYY172 pKa = 10.19FRR174 pKa = 11.84SPQLALSSGPEE185 pKa = 3.72SVMATRR191 pKa = 11.84CSLQIRR197 pKa = 11.84NITQVLNQGGVVRR210 pKa = 11.84SLRR213 pKa = 11.84VTTGIINPGIVNGPTQDD230 pKa = 4.15LLDD233 pKa = 4.2LAEE236 pKa = 4.38HH237 pKa = 5.92VRR239 pKa = 11.84NHH241 pKa = 5.33ARR243 pKa = 11.84TRR245 pKa = 11.84TYY247 pKa = 11.03AGADD251 pKa = 3.22LVEE254 pKa = 4.47NKK256 pKa = 9.88QINCTVVDD264 pKa = 3.76QSRR267 pKa = 11.84ATTFLDD273 pKa = 3.77FGLQVPVEE281 pKa = 4.35DD282 pKa = 5.27LPWIPPGSGVSVGPYY297 pKa = 9.54TLGLHH302 pKa = 6.07NPTYY306 pKa = 10.54TPIILLFEE314 pKa = 4.51SFSTTNEE321 pKa = 3.7YY322 pKa = 10.47EE323 pKa = 3.71ISIRR327 pKa = 11.84TQFLAHH333 pKa = 5.8YY334 pKa = 8.64VQGSMLANLAITPPSVGDD352 pKa = 3.51KK353 pKa = 11.21LNVHH357 pKa = 7.06RR358 pKa = 11.84DD359 pKa = 3.11NEE361 pKa = 4.11EE362 pKa = 3.76RR363 pKa = 11.84KK364 pKa = 10.44GSVLHH369 pKa = 6.61DD370 pKa = 3.2VSVALKK376 pKa = 10.49HH377 pKa = 5.8AGNWAMEE384 pKa = 4.25HH385 pKa = 6.39GGEE388 pKa = 4.03IMGGIGAAKK397 pKa = 8.73WAVPRR402 pKa = 11.84VGQALGWGAKK412 pKa = 8.94LAPLLL417 pKa = 4.31
MM1 pKa = 7.72ALTAKK6 pKa = 7.91QQRR9 pKa = 11.84AIADD13 pKa = 3.55AAPAAKK19 pKa = 10.06AAMRR23 pKa = 11.84AAFALQNRR31 pKa = 11.84GRR33 pKa = 11.84VPGVDD38 pKa = 3.35RR39 pKa = 11.84NAKK42 pKa = 6.97QARR45 pKa = 11.84ARR47 pKa = 11.84GRR49 pKa = 11.84RR50 pKa = 11.84GIPTAIQNMPRR61 pKa = 11.84GAAGPRR67 pKa = 11.84GEE69 pKa = 4.8MPDD72 pKa = 2.99ATRR75 pKa = 11.84FHH77 pKa = 7.45WDD79 pKa = 3.11QTPASSNVVAPRR91 pKa = 11.84GFGYY95 pKa = 10.95YY96 pKa = 10.19DD97 pKa = 4.28AFTHH101 pKa = 6.05SPSDD105 pKa = 3.23AVTAFSVGPATPIQANTRR123 pKa = 11.84ANVTTSAPDD132 pKa = 3.55GVGAGDD138 pKa = 4.82LGLSMIIVYY147 pKa = 8.74PAATDD152 pKa = 3.39AQAALFKK159 pKa = 10.84CGATATDD166 pKa = 4.4PVTTTYY172 pKa = 10.19FRR174 pKa = 11.84SPQLALSSGPEE185 pKa = 3.72SVMATRR191 pKa = 11.84CSLQIRR197 pKa = 11.84NITQVLNQGGVVRR210 pKa = 11.84SLRR213 pKa = 11.84VTTGIINPGIVNGPTQDD230 pKa = 4.15LLDD233 pKa = 4.2LAEE236 pKa = 4.38HH237 pKa = 5.92VRR239 pKa = 11.84NHH241 pKa = 5.33ARR243 pKa = 11.84TRR245 pKa = 11.84TYY247 pKa = 11.03AGADD251 pKa = 3.22LVEE254 pKa = 4.47NKK256 pKa = 9.88QINCTVVDD264 pKa = 3.76QSRR267 pKa = 11.84ATTFLDD273 pKa = 3.77FGLQVPVEE281 pKa = 4.35DD282 pKa = 5.27LPWIPPGSGVSVGPYY297 pKa = 9.54TLGLHH302 pKa = 6.07NPTYY306 pKa = 10.54TPIILLFEE314 pKa = 4.51SFSTTNEE321 pKa = 3.7YY322 pKa = 10.47EE323 pKa = 3.71ISIRR327 pKa = 11.84TQFLAHH333 pKa = 5.8YY334 pKa = 8.64VQGSMLANLAITPPSVGDD352 pKa = 3.51KK353 pKa = 11.21LNVHH357 pKa = 7.06RR358 pKa = 11.84DD359 pKa = 3.11NEE361 pKa = 4.11EE362 pKa = 3.76RR363 pKa = 11.84KK364 pKa = 10.44GSVLHH369 pKa = 6.61DD370 pKa = 3.2VSVALKK376 pKa = 10.49HH377 pKa = 5.8AGNWAMEE384 pKa = 4.25HH385 pKa = 6.39GGEE388 pKa = 4.03IMGGIGAAKK397 pKa = 8.73WAVPRR402 pKa = 11.84VGQALGWGAKK412 pKa = 8.94LAPLLL417 pKa = 4.31
Molecular weight: 44.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1341 |
417 |
924 |
670.5 |
73.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.545 ± 1.859 | 2.237 ± 0.726 |
5.593 ± 0.496 | 5.444 ± 1.228 |
3.729 ± 0.407 | 8.054 ± 0.622 |
2.535 ± 0.066 | 5.071 ± 0.131 |
4.922 ± 1.208 | 7.159 ± 0.132 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.461 ± 0.145 | 3.43 ± 0.539 |
6.264 ± 0.33 | 3.505 ± 0.503 |
6.562 ± 0.042 | 5.742 ± 0.108 |
6.786 ± 0.54 | 6.786 ± 0.425 |
1.715 ± 0.247 | 2.461 ± 0.145 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
