
Chicory yellow mottle virus large satellite RNA
Taxonomy: Viruses; Satellites; RNA satellites; Single stranded RNA satellites; unclassified Single stranded RNA satellites
Average proteome isoelectric point is 9.16
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
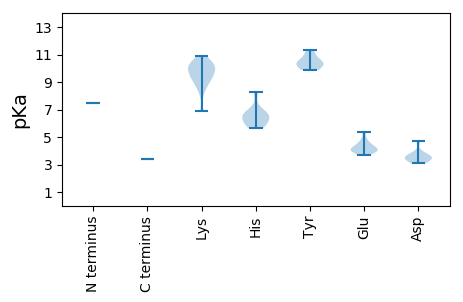
Protein with the lowest isoelectric point:
>tr|Q04325|Q04325_9VIRU Uncharacterized protein OS=Chicory yellow mottle virus large satellite RNA OX=192021 PE=4 SV=1
MM1 pKa = 7.44ARR3 pKa = 11.84TVIIHH8 pKa = 6.6AVASTRR14 pKa = 11.84SVTASPTTANGRR26 pKa = 11.84GRR28 pKa = 11.84ASRR31 pKa = 11.84KK32 pKa = 6.92CTKK35 pKa = 10.48NNAAVEE41 pKa = 4.22SSRR44 pKa = 11.84PRR46 pKa = 11.84KK47 pKa = 9.16LQRR50 pKa = 11.84TAPIKK55 pKa = 10.1TGTRR59 pKa = 11.84DD60 pKa = 3.24NRR62 pKa = 11.84SYY64 pKa = 11.3AQVVMEE70 pKa = 4.57NCHH73 pKa = 6.59HH74 pKa = 5.83SHH76 pKa = 5.66QSKK79 pKa = 10.37ARR81 pKa = 11.84SSEE84 pKa = 3.84KK85 pKa = 10.08FVAAKK90 pKa = 10.01KK91 pKa = 9.21VARR94 pKa = 11.84PSVAVPSKK102 pKa = 10.1LTTGNKK108 pKa = 9.45FDD110 pKa = 3.41VLGFEE115 pKa = 4.15RR116 pKa = 11.84HH117 pKa = 5.95AIDD120 pKa = 3.43RR121 pKa = 11.84AVATSVPTKK130 pKa = 10.36SVQEE134 pKa = 4.15VVSVQKK140 pKa = 9.64GTQYY144 pKa = 10.78VVNHH148 pKa = 6.63TGHH151 pKa = 6.68SYY153 pKa = 9.88GCSRR157 pKa = 11.84KK158 pKa = 10.05GNYY161 pKa = 10.04ASTSYY166 pKa = 10.35RR167 pKa = 11.84CCAKK171 pKa = 10.36VVEE174 pKa = 4.45EE175 pKa = 4.14PVMEE179 pKa = 4.52KK180 pKa = 9.31KK181 pKa = 8.82TVFTDD186 pKa = 3.71VPDD189 pKa = 3.81EE190 pKa = 4.09AAEE193 pKa = 4.09LLRR196 pKa = 11.84QEE198 pKa = 5.39FLAKK202 pKa = 9.6MPQLGHH208 pKa = 6.3SVTLCPVGVDD218 pKa = 3.08TLAWGNCSEE227 pKa = 4.87LDD229 pKa = 3.53SAVAAKK235 pKa = 9.02PAWFGVPVKK244 pKa = 8.31QQRR247 pKa = 11.84KK248 pKa = 8.56KK249 pKa = 9.64MSRR252 pKa = 11.84RR253 pKa = 11.84DD254 pKa = 3.56LRR256 pKa = 11.84NLLSALRR263 pKa = 11.84LARR266 pKa = 11.84TTVNKK271 pKa = 10.41SCSLGDD277 pKa = 3.29AFMWAWRR284 pKa = 11.84TLGFGGAAPLPDD296 pKa = 3.6LKK298 pKa = 10.89GWEE301 pKa = 3.94LSHH304 pKa = 8.28RR305 pKa = 11.84EE306 pKa = 3.72EE307 pKa = 3.99RR308 pKa = 11.84AAAPLHH314 pKa = 6.11MYY316 pKa = 10.39VLVIRR321 pKa = 11.84CGGVGFSLWTGCFDD335 pKa = 4.73APGKK339 pKa = 9.73FIPAMVFHH347 pKa = 6.86GFLLPLSYY355 pKa = 10.4SWQSLEE361 pKa = 3.93VV362 pKa = 3.37
MM1 pKa = 7.44ARR3 pKa = 11.84TVIIHH8 pKa = 6.6AVASTRR14 pKa = 11.84SVTASPTTANGRR26 pKa = 11.84GRR28 pKa = 11.84ASRR31 pKa = 11.84KK32 pKa = 6.92CTKK35 pKa = 10.48NNAAVEE41 pKa = 4.22SSRR44 pKa = 11.84PRR46 pKa = 11.84KK47 pKa = 9.16LQRR50 pKa = 11.84TAPIKK55 pKa = 10.1TGTRR59 pKa = 11.84DD60 pKa = 3.24NRR62 pKa = 11.84SYY64 pKa = 11.3AQVVMEE70 pKa = 4.57NCHH73 pKa = 6.59HH74 pKa = 5.83SHH76 pKa = 5.66QSKK79 pKa = 10.37ARR81 pKa = 11.84SSEE84 pKa = 3.84KK85 pKa = 10.08FVAAKK90 pKa = 10.01KK91 pKa = 9.21VARR94 pKa = 11.84PSVAVPSKK102 pKa = 10.1LTTGNKK108 pKa = 9.45FDD110 pKa = 3.41VLGFEE115 pKa = 4.15RR116 pKa = 11.84HH117 pKa = 5.95AIDD120 pKa = 3.43RR121 pKa = 11.84AVATSVPTKK130 pKa = 10.36SVQEE134 pKa = 4.15VVSVQKK140 pKa = 9.64GTQYY144 pKa = 10.78VVNHH148 pKa = 6.63TGHH151 pKa = 6.68SYY153 pKa = 9.88GCSRR157 pKa = 11.84KK158 pKa = 10.05GNYY161 pKa = 10.04ASTSYY166 pKa = 10.35RR167 pKa = 11.84CCAKK171 pKa = 10.36VVEE174 pKa = 4.45EE175 pKa = 4.14PVMEE179 pKa = 4.52KK180 pKa = 9.31KK181 pKa = 8.82TVFTDD186 pKa = 3.71VPDD189 pKa = 3.81EE190 pKa = 4.09AAEE193 pKa = 4.09LLRR196 pKa = 11.84QEE198 pKa = 5.39FLAKK202 pKa = 9.6MPQLGHH208 pKa = 6.3SVTLCPVGVDD218 pKa = 3.08TLAWGNCSEE227 pKa = 4.87LDD229 pKa = 3.53SAVAAKK235 pKa = 9.02PAWFGVPVKK244 pKa = 8.31QQRR247 pKa = 11.84KK248 pKa = 8.56KK249 pKa = 9.64MSRR252 pKa = 11.84RR253 pKa = 11.84DD254 pKa = 3.56LRR256 pKa = 11.84NLLSALRR263 pKa = 11.84LARR266 pKa = 11.84TTVNKK271 pKa = 10.41SCSLGDD277 pKa = 3.29AFMWAWRR284 pKa = 11.84TLGFGGAAPLPDD296 pKa = 3.6LKK298 pKa = 10.89GWEE301 pKa = 3.94LSHH304 pKa = 8.28RR305 pKa = 11.84EE306 pKa = 3.72EE307 pKa = 3.99RR308 pKa = 11.84AAAPLHH314 pKa = 6.11MYY316 pKa = 10.39VLVIRR321 pKa = 11.84CGGVGFSLWTGCFDD335 pKa = 4.73APGKK339 pKa = 9.73FIPAMVFHH347 pKa = 6.86GFLLPLSYY355 pKa = 10.4SWQSLEE361 pKa = 3.93VV362 pKa = 3.37
Molecular weight: 39.66 kDa
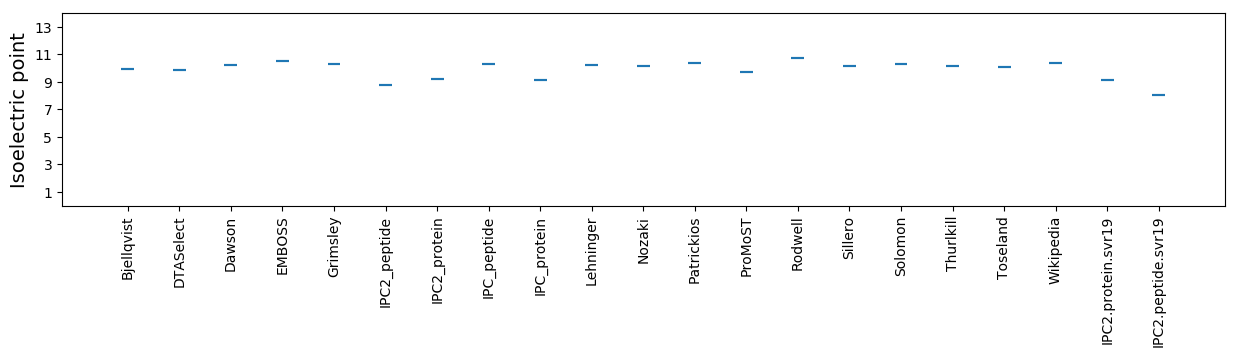
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q04325|Q04325_9VIRU Uncharacterized protein OS=Chicory yellow mottle virus large satellite RNA OX=192021 PE=4 SV=1
MM1 pKa = 7.44ARR3 pKa = 11.84TVIIHH8 pKa = 6.6AVASTRR14 pKa = 11.84SVTASPTTANGRR26 pKa = 11.84GRR28 pKa = 11.84ASRR31 pKa = 11.84KK32 pKa = 6.92CTKK35 pKa = 10.48NNAAVEE41 pKa = 4.22SSRR44 pKa = 11.84PRR46 pKa = 11.84KK47 pKa = 9.16LQRR50 pKa = 11.84TAPIKK55 pKa = 10.1TGTRR59 pKa = 11.84DD60 pKa = 3.24NRR62 pKa = 11.84SYY64 pKa = 11.3AQVVMEE70 pKa = 4.57NCHH73 pKa = 6.59HH74 pKa = 5.83SHH76 pKa = 5.66QSKK79 pKa = 10.37ARR81 pKa = 11.84SSEE84 pKa = 3.84KK85 pKa = 10.08FVAAKK90 pKa = 10.01KK91 pKa = 9.21VARR94 pKa = 11.84PSVAVPSKK102 pKa = 10.1LTTGNKK108 pKa = 9.45FDD110 pKa = 3.41VLGFEE115 pKa = 4.15RR116 pKa = 11.84HH117 pKa = 5.95AIDD120 pKa = 3.43RR121 pKa = 11.84AVATSVPTKK130 pKa = 10.36SVQEE134 pKa = 4.15VVSVQKK140 pKa = 9.64GTQYY144 pKa = 10.78VVNHH148 pKa = 6.63TGHH151 pKa = 6.68SYY153 pKa = 9.88GCSRR157 pKa = 11.84KK158 pKa = 10.05GNYY161 pKa = 10.04ASTSYY166 pKa = 10.35RR167 pKa = 11.84CCAKK171 pKa = 10.36VVEE174 pKa = 4.45EE175 pKa = 4.14PVMEE179 pKa = 4.52KK180 pKa = 9.31KK181 pKa = 8.82TVFTDD186 pKa = 3.71VPDD189 pKa = 3.81EE190 pKa = 4.09AAEE193 pKa = 4.09LLRR196 pKa = 11.84QEE198 pKa = 5.39FLAKK202 pKa = 9.6MPQLGHH208 pKa = 6.3SVTLCPVGVDD218 pKa = 3.08TLAWGNCSEE227 pKa = 4.87LDD229 pKa = 3.53SAVAAKK235 pKa = 9.02PAWFGVPVKK244 pKa = 8.31QQRR247 pKa = 11.84KK248 pKa = 8.56KK249 pKa = 9.64MSRR252 pKa = 11.84RR253 pKa = 11.84DD254 pKa = 3.56LRR256 pKa = 11.84NLLSALRR263 pKa = 11.84LARR266 pKa = 11.84TTVNKK271 pKa = 10.41SCSLGDD277 pKa = 3.29AFMWAWRR284 pKa = 11.84TLGFGGAAPLPDD296 pKa = 3.6LKK298 pKa = 10.89GWEE301 pKa = 3.94LSHH304 pKa = 8.28RR305 pKa = 11.84EE306 pKa = 3.72EE307 pKa = 3.99RR308 pKa = 11.84AAAPLHH314 pKa = 6.11MYY316 pKa = 10.39VLVIRR321 pKa = 11.84CGGVGFSLWTGCFDD335 pKa = 4.73APGKK339 pKa = 9.73FIPAMVFHH347 pKa = 6.86GFLLPLSYY355 pKa = 10.4SWQSLEE361 pKa = 3.93VV362 pKa = 3.37
MM1 pKa = 7.44ARR3 pKa = 11.84TVIIHH8 pKa = 6.6AVASTRR14 pKa = 11.84SVTASPTTANGRR26 pKa = 11.84GRR28 pKa = 11.84ASRR31 pKa = 11.84KK32 pKa = 6.92CTKK35 pKa = 10.48NNAAVEE41 pKa = 4.22SSRR44 pKa = 11.84PRR46 pKa = 11.84KK47 pKa = 9.16LQRR50 pKa = 11.84TAPIKK55 pKa = 10.1TGTRR59 pKa = 11.84DD60 pKa = 3.24NRR62 pKa = 11.84SYY64 pKa = 11.3AQVVMEE70 pKa = 4.57NCHH73 pKa = 6.59HH74 pKa = 5.83SHH76 pKa = 5.66QSKK79 pKa = 10.37ARR81 pKa = 11.84SSEE84 pKa = 3.84KK85 pKa = 10.08FVAAKK90 pKa = 10.01KK91 pKa = 9.21VARR94 pKa = 11.84PSVAVPSKK102 pKa = 10.1LTTGNKK108 pKa = 9.45FDD110 pKa = 3.41VLGFEE115 pKa = 4.15RR116 pKa = 11.84HH117 pKa = 5.95AIDD120 pKa = 3.43RR121 pKa = 11.84AVATSVPTKK130 pKa = 10.36SVQEE134 pKa = 4.15VVSVQKK140 pKa = 9.64GTQYY144 pKa = 10.78VVNHH148 pKa = 6.63TGHH151 pKa = 6.68SYY153 pKa = 9.88GCSRR157 pKa = 11.84KK158 pKa = 10.05GNYY161 pKa = 10.04ASTSYY166 pKa = 10.35RR167 pKa = 11.84CCAKK171 pKa = 10.36VVEE174 pKa = 4.45EE175 pKa = 4.14PVMEE179 pKa = 4.52KK180 pKa = 9.31KK181 pKa = 8.82TVFTDD186 pKa = 3.71VPDD189 pKa = 3.81EE190 pKa = 4.09AAEE193 pKa = 4.09LLRR196 pKa = 11.84QEE198 pKa = 5.39FLAKK202 pKa = 9.6MPQLGHH208 pKa = 6.3SVTLCPVGVDD218 pKa = 3.08TLAWGNCSEE227 pKa = 4.87LDD229 pKa = 3.53SAVAAKK235 pKa = 9.02PAWFGVPVKK244 pKa = 8.31QQRR247 pKa = 11.84KK248 pKa = 8.56KK249 pKa = 9.64MSRR252 pKa = 11.84RR253 pKa = 11.84DD254 pKa = 3.56LRR256 pKa = 11.84NLLSALRR263 pKa = 11.84LARR266 pKa = 11.84TTVNKK271 pKa = 10.41SCSLGDD277 pKa = 3.29AFMWAWRR284 pKa = 11.84TLGFGGAAPLPDD296 pKa = 3.6LKK298 pKa = 10.89GWEE301 pKa = 3.94LSHH304 pKa = 8.28RR305 pKa = 11.84EE306 pKa = 3.72EE307 pKa = 3.99RR308 pKa = 11.84AAAPLHH314 pKa = 6.11MYY316 pKa = 10.39VLVIRR321 pKa = 11.84CGGVGFSLWTGCFDD335 pKa = 4.73APGKK339 pKa = 9.73FIPAMVFHH347 pKa = 6.86GFLLPLSYY355 pKa = 10.4SWQSLEE361 pKa = 3.93VV362 pKa = 3.37
Molecular weight: 39.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
362 |
362 |
362 |
362.0 |
39.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.773 ± 0.0 | 2.762 ± 0.0 |
3.039 ± 0.0 | 4.42 ± 0.0 |
3.591 ± 0.0 | 6.63 ± 0.0 |
3.039 ± 0.0 | 1.657 ± 0.0 |
6.63 ± 0.0 | 7.459 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.21 ± 0.0 | 3.039 ± 0.0 |
4.972 ± 0.0 | 3.039 ± 0.0 |
7.459 ± 0.0 | 8.84 ± 0.0 |
6.63 ± 0.0 | 9.945 ± 0.0 |
1.934 ± 0.0 | 1.934 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
