
Sphingomonas sp. Leaf20
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
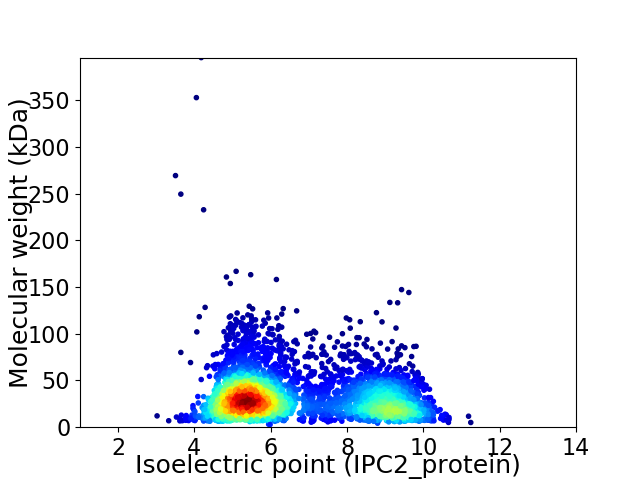
Virtual 2D-PAGE plot for 3757 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
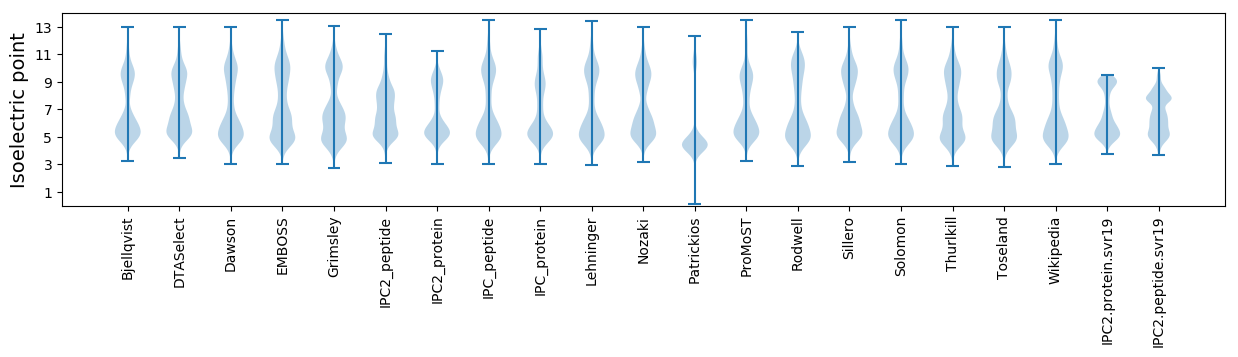
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q4FU37|A0A0Q4FU37_9SPHN Uncharacterized protein OS=Sphingomonas sp. Leaf20 OX=1735685 GN=ASE72_17950 PE=4 SV=1
MM1 pKa = 7.21TFEE4 pKa = 5.06ASGSTAGFNDD14 pKa = 3.33GGTYY18 pKa = 10.47NDD20 pKa = 4.46GGPVPLAASLDD31 pKa = 4.02PNAGGTSNDD40 pKa = 3.15KK41 pKa = 10.4TIWTLDD47 pKa = 3.4QINANFNRR55 pKa = 11.84SGYY58 pKa = 10.41DD59 pKa = 2.68WYY61 pKa = 9.94TNNYY65 pKa = 9.53GEE67 pKa = 5.3LNDD70 pKa = 4.33GVLNYY75 pKa = 9.97GFWLNQEE82 pKa = 4.28EE83 pKa = 4.92LGNSYY88 pKa = 8.45YY89 pKa = 10.99TNVTGTVALNEE100 pKa = 4.25YY101 pKa = 10.9FDD103 pKa = 3.89FTAFNAGQTALAQRR117 pKa = 11.84SMTLWDD123 pKa = 4.19DD124 pKa = 3.72LVAISFEE131 pKa = 4.23QTKK134 pKa = 10.63SGSADD139 pKa = 2.74ITFGNTATGGAQAYY153 pKa = 9.58AYY155 pKa = 10.36LPFGSTDD162 pKa = 3.49DD163 pKa = 4.51AEE165 pKa = 4.25ILADD169 pKa = 5.76FGFEE173 pKa = 4.23DD174 pKa = 4.19YY175 pKa = 11.3GRR177 pKa = 11.84LGGDD181 pKa = 3.02VWIDD185 pKa = 3.13GGVASNFSPLTASYY199 pKa = 10.05YY200 pKa = 10.34AQTTMVHH207 pKa = 6.7EE208 pKa = 4.89IGHH211 pKa = 7.04AIGLSHH217 pKa = 7.4PGDD220 pKa = 4.1YY221 pKa = 10.71NALDD225 pKa = 4.91DD226 pKa = 4.39NDD228 pKa = 4.33GDD230 pKa = 4.39GVPDD234 pKa = 5.79PITYY238 pKa = 10.63ASDD241 pKa = 2.91AFYY244 pKa = 10.88AQDD247 pKa = 3.19SRR249 pKa = 11.84QYY251 pKa = 11.43SIMSYY256 pKa = 10.4FDD258 pKa = 3.68AYY260 pKa = 9.1EE261 pKa = 4.12TGAQHH266 pKa = 7.31IDD268 pKa = 2.86WALLNFAYY276 pKa = 10.26AATPLVHH283 pKa = 7.61DD284 pKa = 4.55IAAIQKK290 pKa = 9.94IYY292 pKa = 10.77GVDD295 pKa = 3.03QTTRR299 pKa = 11.84TGNTVYY305 pKa = 10.74GFNATADD312 pKa = 3.52NAVYY316 pKa = 10.46DD317 pKa = 5.35FSANTRR323 pKa = 11.84PIVAIWDD330 pKa = 3.59AGGTDD335 pKa = 3.56TLDD338 pKa = 3.36FSGWNTSSTIDD349 pKa = 3.7LNQGAFSSGGGVDD362 pKa = 5.22DD363 pKa = 5.7FLTLEE368 pKa = 3.97QVNANRR374 pKa = 11.84AALGFAARR382 pKa = 11.84SQASYY387 pKa = 11.58DD388 pKa = 3.58FYY390 pKa = 11.72EE391 pKa = 4.59DD392 pKa = 4.86LKK394 pKa = 10.03TQLGLKK400 pKa = 10.19SGLFTDD406 pKa = 4.37NVSIAYY412 pKa = 9.4GAVIEE417 pKa = 4.08NAIGGGGNDD426 pKa = 3.53RR427 pKa = 11.84IIGNEE432 pKa = 3.84VANALTGGAGRR443 pKa = 11.84DD444 pKa = 3.25IFEE447 pKa = 4.73LRR449 pKa = 11.84SGGTSGADD457 pKa = 4.17RR458 pKa = 11.84ITDD461 pKa = 3.58WSRR464 pKa = 11.84GDD466 pKa = 3.43VLATTKK472 pKa = 10.69AINDD476 pKa = 3.74ANGDD480 pKa = 4.37GIITWKK486 pKa = 10.44GASLTLDD493 pKa = 3.45GSDD496 pKa = 3.53GDD498 pKa = 3.82RR499 pKa = 11.84VEE501 pKa = 5.12LVGAQGSSGLRR512 pKa = 11.84LMGSIDD518 pKa = 3.34GVFYY522 pKa = 11.46YY523 pKa = 10.62EE524 pKa = 4.33DD525 pKa = 3.38ARR527 pKa = 11.84VRR529 pKa = 11.84PVASSGQKK537 pKa = 9.65VYY539 pKa = 11.11EE540 pKa = 4.26SGFGNDD546 pKa = 3.26VLRR549 pKa = 11.84GRR551 pKa = 11.84TTASATDD558 pKa = 3.3VFFFDD563 pKa = 3.51TANPVSSLGKK573 pKa = 9.18DD574 pKa = 3.0TVSFTKK580 pKa = 10.6NDD582 pKa = 3.08IFVTTTKK589 pKa = 10.75LADD592 pKa = 3.63ANNDD596 pKa = 3.11NKK598 pKa = 9.78ITFGKK603 pKa = 10.4NGVLDD608 pKa = 4.03LGGDD612 pKa = 3.6HH613 pKa = 7.09GSVALSGTSALEE625 pKa = 3.69YY626 pKa = 11.0DD627 pKa = 4.16GVVTNNGTEE636 pKa = 4.15YY637 pKa = 10.73YY638 pKa = 10.15VYY640 pKa = 10.93SNVGSAVGVGALHH653 pKa = 6.88FF654 pKa = 4.51
MM1 pKa = 7.21TFEE4 pKa = 5.06ASGSTAGFNDD14 pKa = 3.33GGTYY18 pKa = 10.47NDD20 pKa = 4.46GGPVPLAASLDD31 pKa = 4.02PNAGGTSNDD40 pKa = 3.15KK41 pKa = 10.4TIWTLDD47 pKa = 3.4QINANFNRR55 pKa = 11.84SGYY58 pKa = 10.41DD59 pKa = 2.68WYY61 pKa = 9.94TNNYY65 pKa = 9.53GEE67 pKa = 5.3LNDD70 pKa = 4.33GVLNYY75 pKa = 9.97GFWLNQEE82 pKa = 4.28EE83 pKa = 4.92LGNSYY88 pKa = 8.45YY89 pKa = 10.99TNVTGTVALNEE100 pKa = 4.25YY101 pKa = 10.9FDD103 pKa = 3.89FTAFNAGQTALAQRR117 pKa = 11.84SMTLWDD123 pKa = 4.19DD124 pKa = 3.72LVAISFEE131 pKa = 4.23QTKK134 pKa = 10.63SGSADD139 pKa = 2.74ITFGNTATGGAQAYY153 pKa = 9.58AYY155 pKa = 10.36LPFGSTDD162 pKa = 3.49DD163 pKa = 4.51AEE165 pKa = 4.25ILADD169 pKa = 5.76FGFEE173 pKa = 4.23DD174 pKa = 4.19YY175 pKa = 11.3GRR177 pKa = 11.84LGGDD181 pKa = 3.02VWIDD185 pKa = 3.13GGVASNFSPLTASYY199 pKa = 10.05YY200 pKa = 10.34AQTTMVHH207 pKa = 6.7EE208 pKa = 4.89IGHH211 pKa = 7.04AIGLSHH217 pKa = 7.4PGDD220 pKa = 4.1YY221 pKa = 10.71NALDD225 pKa = 4.91DD226 pKa = 4.39NDD228 pKa = 4.33GDD230 pKa = 4.39GVPDD234 pKa = 5.79PITYY238 pKa = 10.63ASDD241 pKa = 2.91AFYY244 pKa = 10.88AQDD247 pKa = 3.19SRR249 pKa = 11.84QYY251 pKa = 11.43SIMSYY256 pKa = 10.4FDD258 pKa = 3.68AYY260 pKa = 9.1EE261 pKa = 4.12TGAQHH266 pKa = 7.31IDD268 pKa = 2.86WALLNFAYY276 pKa = 10.26AATPLVHH283 pKa = 7.61DD284 pKa = 4.55IAAIQKK290 pKa = 9.94IYY292 pKa = 10.77GVDD295 pKa = 3.03QTTRR299 pKa = 11.84TGNTVYY305 pKa = 10.74GFNATADD312 pKa = 3.52NAVYY316 pKa = 10.46DD317 pKa = 5.35FSANTRR323 pKa = 11.84PIVAIWDD330 pKa = 3.59AGGTDD335 pKa = 3.56TLDD338 pKa = 3.36FSGWNTSSTIDD349 pKa = 3.7LNQGAFSSGGGVDD362 pKa = 5.22DD363 pKa = 5.7FLTLEE368 pKa = 3.97QVNANRR374 pKa = 11.84AALGFAARR382 pKa = 11.84SQASYY387 pKa = 11.58DD388 pKa = 3.58FYY390 pKa = 11.72EE391 pKa = 4.59DD392 pKa = 4.86LKK394 pKa = 10.03TQLGLKK400 pKa = 10.19SGLFTDD406 pKa = 4.37NVSIAYY412 pKa = 9.4GAVIEE417 pKa = 4.08NAIGGGGNDD426 pKa = 3.53RR427 pKa = 11.84IIGNEE432 pKa = 3.84VANALTGGAGRR443 pKa = 11.84DD444 pKa = 3.25IFEE447 pKa = 4.73LRR449 pKa = 11.84SGGTSGADD457 pKa = 4.17RR458 pKa = 11.84ITDD461 pKa = 3.58WSRR464 pKa = 11.84GDD466 pKa = 3.43VLATTKK472 pKa = 10.69AINDD476 pKa = 3.74ANGDD480 pKa = 4.37GIITWKK486 pKa = 10.44GASLTLDD493 pKa = 3.45GSDD496 pKa = 3.53GDD498 pKa = 3.82RR499 pKa = 11.84VEE501 pKa = 5.12LVGAQGSSGLRR512 pKa = 11.84LMGSIDD518 pKa = 3.34GVFYY522 pKa = 11.46YY523 pKa = 10.62EE524 pKa = 4.33DD525 pKa = 3.38ARR527 pKa = 11.84VRR529 pKa = 11.84PVASSGQKK537 pKa = 9.65VYY539 pKa = 11.11EE540 pKa = 4.26SGFGNDD546 pKa = 3.26VLRR549 pKa = 11.84GRR551 pKa = 11.84TTASATDD558 pKa = 3.3VFFFDD563 pKa = 3.51TANPVSSLGKK573 pKa = 9.18DD574 pKa = 3.0TVSFTKK580 pKa = 10.6NDD582 pKa = 3.08IFVTTTKK589 pKa = 10.75LADD592 pKa = 3.63ANNDD596 pKa = 3.11NKK598 pKa = 9.78ITFGKK603 pKa = 10.4NGVLDD608 pKa = 4.03LGGDD612 pKa = 3.6HH613 pKa = 7.09GSVALSGTSALEE625 pKa = 3.69YY626 pKa = 11.0DD627 pKa = 4.16GVVTNNGTEE636 pKa = 4.15YY637 pKa = 10.73YY638 pKa = 10.15VYY640 pKa = 10.93SNVGSAVGVGALHH653 pKa = 6.88FF654 pKa = 4.51
Molecular weight: 69.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q4GBT3|A0A0Q4GBT3_9SPHN 30S ribosomal protein S7 OS=Sphingomonas sp. Leaf20 OX=1735685 GN=rpsG PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVIHH32 pKa = 6.17ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVIHH32 pKa = 6.17ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1227095 |
26 |
4130 |
326.6 |
35.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.881 ± 0.067 | 0.707 ± 0.011 |
6.227 ± 0.035 | 4.665 ± 0.042 |
3.435 ± 0.022 | 9.088 ± 0.06 |
1.894 ± 0.021 | 5.081 ± 0.03 |
2.687 ± 0.033 | 9.588 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.366 ± 0.022 | 2.466 ± 0.039 |
5.283 ± 0.034 | 2.977 ± 0.024 |
7.398 ± 0.051 | 5.143 ± 0.034 |
5.994 ± 0.042 | 7.537 ± 0.032 |
1.363 ± 0.018 | 2.219 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
