
Hubei picorna-like virus 75
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
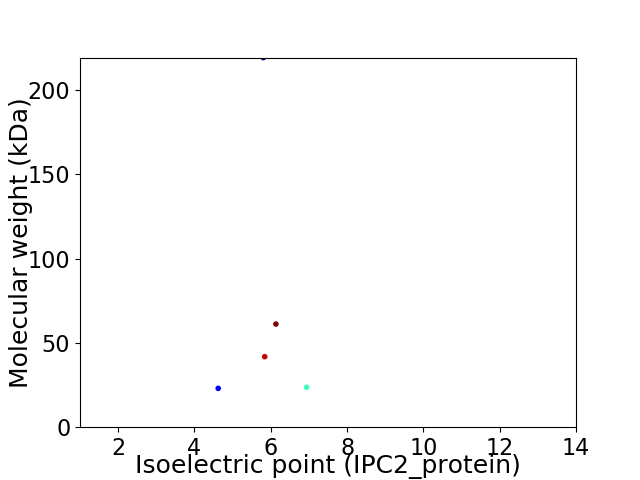
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
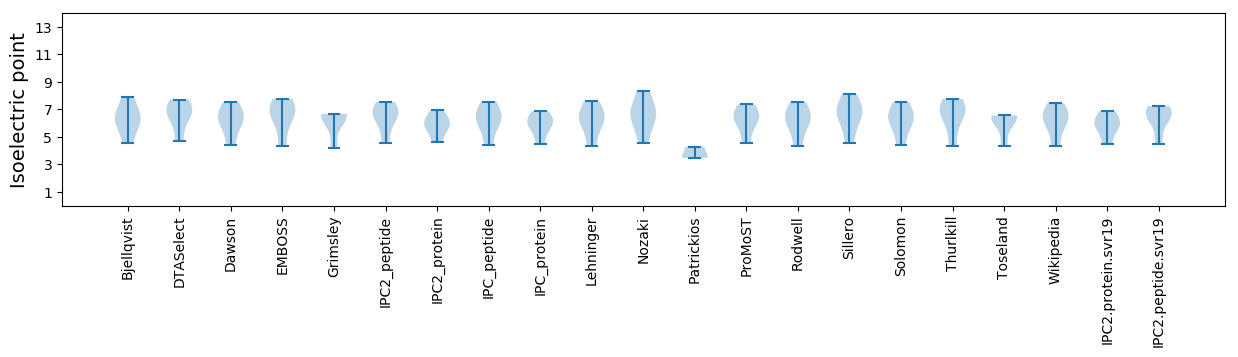
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLU5|A0A1L3KLU5_9VIRU Calici_coat domain-containing protein OS=Hubei picorna-like virus 75 OX=1923159 PE=4 SV=1
MM1 pKa = 7.6ASIAMGIGAVVSAIGTAASVGTSIANQKK29 pKa = 9.21SANSTTLEE37 pKa = 4.03NQNSVNVTNIQLQHH51 pKa = 6.36DD52 pKa = 3.72QQNFNEE58 pKa = 4.57HH59 pKa = 6.15MFDD62 pKa = 3.83KK63 pKa = 11.06NVDD66 pKa = 3.57LFTQAGLPGYY76 pKa = 8.25MAAMGGGMNNPLYY89 pKa = 10.66DD90 pKa = 3.93AFPRR94 pKa = 11.84TSQQVAGNNFSAMSMPGNVMSEE116 pKa = 3.99YY117 pKa = 10.2GGNVIQQMTGMANYY131 pKa = 9.79RR132 pKa = 11.84VSQRR136 pKa = 11.84SNINPTSTGTQTPSGSGTMNPRR158 pKa = 11.84STFSTQTEE166 pKa = 4.66SNPSGTTMYY175 pKa = 7.84ATPNTPQQLWNQLGGTTPSTSQTVAPTAAVAAPAGGSAMPMEE217 pKa = 5.63DD218 pKa = 3.96EE219 pKa = 5.21DD220 pKa = 4.95SSS222 pKa = 3.78
MM1 pKa = 7.6ASIAMGIGAVVSAIGTAASVGTSIANQKK29 pKa = 9.21SANSTTLEE37 pKa = 4.03NQNSVNVTNIQLQHH51 pKa = 6.36DD52 pKa = 3.72QQNFNEE58 pKa = 4.57HH59 pKa = 6.15MFDD62 pKa = 3.83KK63 pKa = 11.06NVDD66 pKa = 3.57LFTQAGLPGYY76 pKa = 8.25MAAMGGGMNNPLYY89 pKa = 10.66DD90 pKa = 3.93AFPRR94 pKa = 11.84TSQQVAGNNFSAMSMPGNVMSEE116 pKa = 3.99YY117 pKa = 10.2GGNVIQQMTGMANYY131 pKa = 9.79RR132 pKa = 11.84VSQRR136 pKa = 11.84SNINPTSTGTQTPSGSGTMNPRR158 pKa = 11.84STFSTQTEE166 pKa = 4.66SNPSGTTMYY175 pKa = 7.84ATPNTPQQLWNQLGGTTPSTSQTVAPTAAVAAPAGGSAMPMEE217 pKa = 5.63DD218 pKa = 3.96EE219 pKa = 5.21DD220 pKa = 4.95SSS222 pKa = 3.78
Molecular weight: 23.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KM10|A0A1L3KM10_9VIRU Uncharacterized protein OS=Hubei picorna-like virus 75 OX=1923159 PE=4 SV=1
MM1 pKa = 7.36SLKK4 pKa = 10.47DD5 pKa = 3.44LRR7 pKa = 11.84IWLDD11 pKa = 3.77LIKK14 pKa = 10.74FLDD17 pKa = 3.8NNTEE21 pKa = 3.76IFKK24 pKa = 10.93DD25 pKa = 4.44GSAPEE30 pKa = 4.21RR31 pKa = 11.84PRR33 pKa = 11.84LWTLTRR39 pKa = 11.84QACFGQKK46 pKa = 8.63RR47 pKa = 11.84TWEE50 pKa = 3.94IPEE53 pKa = 3.82VCRR56 pKa = 11.84LIGCFNSTLSTYY68 pKa = 9.65IEE70 pKa = 3.92QMRR73 pKa = 11.84GIGIAIGTRR82 pKa = 11.84GVYY85 pKa = 8.33ITSPRR90 pKa = 11.84YY91 pKa = 7.85PTEE94 pKa = 3.88SDD96 pKa = 3.63LAILAALAPFSCDD109 pKa = 2.85EE110 pKa = 4.33LSKK113 pKa = 9.98HH114 pKa = 5.44TNEE117 pKa = 4.1EE118 pKa = 3.71LCEE121 pKa = 4.17FFTTLYY127 pKa = 10.9GAFEE131 pKa = 4.45NSVSIRR137 pKa = 11.84PKK139 pKa = 10.36LFKK142 pKa = 10.93LFDD145 pKa = 3.98GLRR148 pKa = 11.84HH149 pKa = 5.21FHH151 pKa = 6.79SSTSHH156 pKa = 6.06SSKK159 pKa = 10.3CANAVGKK166 pKa = 9.7FKK168 pKa = 10.65QICNCPHH175 pKa = 6.62SDD177 pKa = 3.74DD178 pKa = 4.93VIRR181 pKa = 11.84SCCSICHH188 pKa = 6.38GDD190 pKa = 3.24VHH192 pKa = 7.54SRR194 pKa = 11.84NTRR197 pKa = 11.84VYY199 pKa = 10.4SSCCSEE205 pKa = 4.21SCTSSS210 pKa = 2.81
MM1 pKa = 7.36SLKK4 pKa = 10.47DD5 pKa = 3.44LRR7 pKa = 11.84IWLDD11 pKa = 3.77LIKK14 pKa = 10.74FLDD17 pKa = 3.8NNTEE21 pKa = 3.76IFKK24 pKa = 10.93DD25 pKa = 4.44GSAPEE30 pKa = 4.21RR31 pKa = 11.84PRR33 pKa = 11.84LWTLTRR39 pKa = 11.84QACFGQKK46 pKa = 8.63RR47 pKa = 11.84TWEE50 pKa = 3.94IPEE53 pKa = 3.82VCRR56 pKa = 11.84LIGCFNSTLSTYY68 pKa = 9.65IEE70 pKa = 3.92QMRR73 pKa = 11.84GIGIAIGTRR82 pKa = 11.84GVYY85 pKa = 8.33ITSPRR90 pKa = 11.84YY91 pKa = 7.85PTEE94 pKa = 3.88SDD96 pKa = 3.63LAILAALAPFSCDD109 pKa = 2.85EE110 pKa = 4.33LSKK113 pKa = 9.98HH114 pKa = 5.44TNEE117 pKa = 4.1EE118 pKa = 3.71LCEE121 pKa = 4.17FFTTLYY127 pKa = 10.9GAFEE131 pKa = 4.45NSVSIRR137 pKa = 11.84PKK139 pKa = 10.36LFKK142 pKa = 10.93LFDD145 pKa = 3.98GLRR148 pKa = 11.84HH149 pKa = 5.21FHH151 pKa = 6.79SSTSHH156 pKa = 6.06SSKK159 pKa = 10.3CANAVGKK166 pKa = 9.7FKK168 pKa = 10.65QICNCPHH175 pKa = 6.62SDD177 pKa = 3.74DD178 pKa = 4.93VIRR181 pKa = 11.84SCCSICHH188 pKa = 6.38GDD190 pKa = 3.24VHH192 pKa = 7.54SRR194 pKa = 11.84NTRR197 pKa = 11.84VYY199 pKa = 10.4SSCCSEE205 pKa = 4.21SCTSSS210 pKa = 2.81
Molecular weight: 23.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3290 |
210 |
1932 |
658.0 |
73.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.565 ± 0.602 | 2.036 ± 0.491 |
5.258 ± 0.448 | 5.502 ± 1.13 |
4.559 ± 0.233 | 5.897 ± 0.634 |
2.492 ± 0.503 | 5.684 ± 0.482 |
5.38 ± 1.184 | 8.298 ± 0.778 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.313 ± 0.52 | 4.924 ± 0.705 |
5.41 ± 0.628 | 4.134 ± 0.553 |
4.347 ± 0.428 | 7.508 ± 1.014 |
7.264 ± 0.638 | 6.565 ± 0.587 |
1.581 ± 0.371 | 3.283 ± 0.415 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
