
Methyloceanibacter caenitepidi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Hyphomicrobiaceae; Methyloceanibacter
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
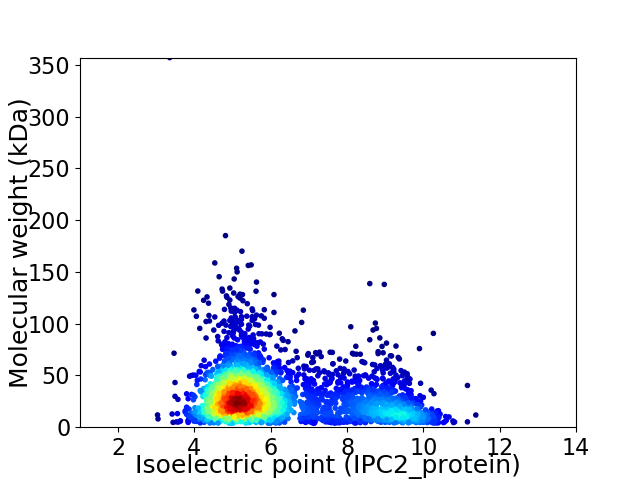
Virtual 2D-PAGE plot for 3341 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A8K2A1|A0A0A8K2A1_9RHIZ ABC transporter domain-containing protein OS=Methyloceanibacter caenitepidi OX=1384459 GN=GL4_0671 PE=3 SV=1
MM1 pKa = 7.43KK2 pKa = 9.04HH3 pKa = 4.33TAYY6 pKa = 10.48VGSISTGTLLTEE18 pKa = 4.73DD19 pKa = 4.04LLEE22 pKa = 4.46AFVGEE27 pKa = 4.58LSFLADD33 pKa = 3.84SVDD36 pKa = 3.63HH37 pKa = 6.71PGNRR41 pKa = 11.84QTYY44 pKa = 10.2EE45 pKa = 4.3DD46 pKa = 4.37LCQEE50 pKa = 5.16AIDD53 pKa = 4.81ADD55 pKa = 4.33PEE57 pKa = 4.32SEE59 pKa = 4.23DD60 pKa = 3.51AQEE63 pKa = 4.46ILNSLIDD70 pKa = 4.27ALTEE74 pKa = 3.81LAPPYY79 pKa = 9.96CYY81 pKa = 10.26FGARR85 pKa = 11.84EE86 pKa = 3.89GDD88 pKa = 3.66GADD91 pKa = 3.22FGFWPDD97 pKa = 3.01TDD99 pKa = 5.09SIAEE103 pKa = 4.27LPRR106 pKa = 11.84VSDD109 pKa = 3.93PNEE112 pKa = 3.7VRR114 pKa = 11.84IADD117 pKa = 3.97HH118 pKa = 7.39DD119 pKa = 3.91WLFVNDD125 pKa = 4.6HH126 pKa = 6.22GNITIYY132 pKa = 10.59SGATAQPILEE142 pKa = 4.36LVV144 pKa = 3.37
MM1 pKa = 7.43KK2 pKa = 9.04HH3 pKa = 4.33TAYY6 pKa = 10.48VGSISTGTLLTEE18 pKa = 4.73DD19 pKa = 4.04LLEE22 pKa = 4.46AFVGEE27 pKa = 4.58LSFLADD33 pKa = 3.84SVDD36 pKa = 3.63HH37 pKa = 6.71PGNRR41 pKa = 11.84QTYY44 pKa = 10.2EE45 pKa = 4.3DD46 pKa = 4.37LCQEE50 pKa = 5.16AIDD53 pKa = 4.81ADD55 pKa = 4.33PEE57 pKa = 4.32SEE59 pKa = 4.23DD60 pKa = 3.51AQEE63 pKa = 4.46ILNSLIDD70 pKa = 4.27ALTEE74 pKa = 3.81LAPPYY79 pKa = 9.96CYY81 pKa = 10.26FGARR85 pKa = 11.84EE86 pKa = 3.89GDD88 pKa = 3.66GADD91 pKa = 3.22FGFWPDD97 pKa = 3.01TDD99 pKa = 5.09SIAEE103 pKa = 4.27LPRR106 pKa = 11.84VSDD109 pKa = 3.93PNEE112 pKa = 3.7VRR114 pKa = 11.84IADD117 pKa = 3.97HH118 pKa = 7.39DD119 pKa = 3.91WLFVNDD125 pKa = 4.6HH126 pKa = 6.22GNITIYY132 pKa = 10.59SGATAQPILEE142 pKa = 4.36LVV144 pKa = 3.37
Molecular weight: 15.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A8K5B1|A0A0A8K5B1_9RHIZ 50S ribosomal protein L25 OS=Methyloceanibacter caenitepidi OX=1384459 GN=rplY PE=3 SV=1
MM1 pKa = 7.39ATRR4 pKa = 11.84TTGKK8 pKa = 10.15KK9 pKa = 8.29RR10 pKa = 11.84TASKK14 pKa = 8.76KK15 pKa = 7.73TPARR19 pKa = 11.84RR20 pKa = 11.84TAARR24 pKa = 11.84RR25 pKa = 11.84TAARR29 pKa = 11.84RR30 pKa = 11.84PAARR34 pKa = 11.84KK35 pKa = 6.38TAARR39 pKa = 11.84KK40 pKa = 6.43TAARR44 pKa = 11.84KK45 pKa = 4.82TTARR49 pKa = 11.84KK50 pKa = 6.62TAARR54 pKa = 11.84KK55 pKa = 6.46TARR58 pKa = 11.84KK59 pKa = 7.64STARR63 pKa = 11.84KK64 pKa = 7.24STARR68 pKa = 11.84KK69 pKa = 6.32TAARR73 pKa = 11.84KK74 pKa = 9.6SPARR78 pKa = 11.84KK79 pKa = 6.81TAARR83 pKa = 11.84KK84 pKa = 6.46TAARR88 pKa = 11.84KK89 pKa = 6.46TARR92 pKa = 11.84KK93 pKa = 7.69STARR97 pKa = 11.84KK98 pKa = 8.3SAARR102 pKa = 11.84KK103 pKa = 6.18TAARR107 pKa = 11.84KK108 pKa = 9.6SPARR112 pKa = 11.84KK113 pKa = 6.81TAARR117 pKa = 11.84KK118 pKa = 6.46TAARR122 pKa = 11.84KK123 pKa = 6.46TARR126 pKa = 11.84KK127 pKa = 7.69STARR131 pKa = 11.84KK132 pKa = 8.3SAARR136 pKa = 11.84KK137 pKa = 6.19TAARR141 pKa = 11.84RR142 pKa = 11.84SPARR146 pKa = 11.84KK147 pKa = 6.76TAARR151 pKa = 11.84KK152 pKa = 6.44TAARR156 pKa = 11.84KK157 pKa = 7.78STARR161 pKa = 11.84KK162 pKa = 7.28STARR166 pKa = 11.84KK167 pKa = 8.27SAARR171 pKa = 11.84KK172 pKa = 7.34STARR176 pKa = 11.84KK177 pKa = 7.28STARR181 pKa = 11.84KK182 pKa = 8.04SAARR186 pKa = 11.84RR187 pKa = 11.84GTARR191 pKa = 11.84KK192 pKa = 9.15SAAKK196 pKa = 10.04KK197 pKa = 9.83SAAKK201 pKa = 10.16KK202 pKa = 9.01SARR205 pKa = 11.84KK206 pKa = 9.62ASARR210 pKa = 11.84KK211 pKa = 9.43APAKK215 pKa = 10.3KK216 pKa = 8.9RR217 pKa = 11.84TAAKK221 pKa = 10.05KK222 pKa = 8.71KK223 pKa = 7.61TAARR227 pKa = 11.84KK228 pKa = 8.37PAVKK232 pKa = 10.26KK233 pKa = 10.5SAAKK237 pKa = 10.24KK238 pKa = 8.48KK239 pKa = 7.79TAARR243 pKa = 11.84KK244 pKa = 6.69TAAKK248 pKa = 8.72KK249 pKa = 8.54TVAKK253 pKa = 10.1KK254 pKa = 9.2KK255 pKa = 8.77TVAKK259 pKa = 10.17KK260 pKa = 9.94AAPKK264 pKa = 9.94KK265 pKa = 10.07AAAKK269 pKa = 9.86KK270 pKa = 9.67AAPKK274 pKa = 9.99KK275 pKa = 9.89AAPKK279 pKa = 9.63KK280 pKa = 9.25AAPKK284 pKa = 9.99KK285 pKa = 10.0VEE287 pKa = 4.22TKK289 pKa = 10.18KK290 pKa = 10.91VEE292 pKa = 4.31PKK294 pKa = 9.78TVAPTPKK301 pKa = 9.53PVASPAPRR309 pKa = 11.84PPVAQPAQRR318 pKa = 11.84PPLAQPSPSPARR330 pKa = 11.84PSSPGAGGTPPRR342 pKa = 11.84HH343 pKa = 5.71VSPGSAIPGYY353 pKa = 9.48QPPRR357 pKa = 11.84PPMPGSTPGPTPTPMPYY374 pKa = 9.6RR375 pKa = 11.84PSWARR380 pKa = 11.84DD381 pKa = 3.24DD382 pKa = 4.42DD383 pKa = 4.0
MM1 pKa = 7.39ATRR4 pKa = 11.84TTGKK8 pKa = 10.15KK9 pKa = 8.29RR10 pKa = 11.84TASKK14 pKa = 8.76KK15 pKa = 7.73TPARR19 pKa = 11.84RR20 pKa = 11.84TAARR24 pKa = 11.84RR25 pKa = 11.84TAARR29 pKa = 11.84RR30 pKa = 11.84PAARR34 pKa = 11.84KK35 pKa = 6.38TAARR39 pKa = 11.84KK40 pKa = 6.43TAARR44 pKa = 11.84KK45 pKa = 4.82TTARR49 pKa = 11.84KK50 pKa = 6.62TAARR54 pKa = 11.84KK55 pKa = 6.46TARR58 pKa = 11.84KK59 pKa = 7.64STARR63 pKa = 11.84KK64 pKa = 7.24STARR68 pKa = 11.84KK69 pKa = 6.32TAARR73 pKa = 11.84KK74 pKa = 9.6SPARR78 pKa = 11.84KK79 pKa = 6.81TAARR83 pKa = 11.84KK84 pKa = 6.46TAARR88 pKa = 11.84KK89 pKa = 6.46TARR92 pKa = 11.84KK93 pKa = 7.69STARR97 pKa = 11.84KK98 pKa = 8.3SAARR102 pKa = 11.84KK103 pKa = 6.18TAARR107 pKa = 11.84KK108 pKa = 9.6SPARR112 pKa = 11.84KK113 pKa = 6.81TAARR117 pKa = 11.84KK118 pKa = 6.46TAARR122 pKa = 11.84KK123 pKa = 6.46TARR126 pKa = 11.84KK127 pKa = 7.69STARR131 pKa = 11.84KK132 pKa = 8.3SAARR136 pKa = 11.84KK137 pKa = 6.19TAARR141 pKa = 11.84RR142 pKa = 11.84SPARR146 pKa = 11.84KK147 pKa = 6.76TAARR151 pKa = 11.84KK152 pKa = 6.44TAARR156 pKa = 11.84KK157 pKa = 7.78STARR161 pKa = 11.84KK162 pKa = 7.28STARR166 pKa = 11.84KK167 pKa = 8.27SAARR171 pKa = 11.84KK172 pKa = 7.34STARR176 pKa = 11.84KK177 pKa = 7.28STARR181 pKa = 11.84KK182 pKa = 8.04SAARR186 pKa = 11.84RR187 pKa = 11.84GTARR191 pKa = 11.84KK192 pKa = 9.15SAAKK196 pKa = 10.04KK197 pKa = 9.83SAAKK201 pKa = 10.16KK202 pKa = 9.01SARR205 pKa = 11.84KK206 pKa = 9.62ASARR210 pKa = 11.84KK211 pKa = 9.43APAKK215 pKa = 10.3KK216 pKa = 8.9RR217 pKa = 11.84TAAKK221 pKa = 10.05KK222 pKa = 8.71KK223 pKa = 7.61TAARR227 pKa = 11.84KK228 pKa = 8.37PAVKK232 pKa = 10.26KK233 pKa = 10.5SAAKK237 pKa = 10.24KK238 pKa = 8.48KK239 pKa = 7.79TAARR243 pKa = 11.84KK244 pKa = 6.69TAAKK248 pKa = 8.72KK249 pKa = 8.54TVAKK253 pKa = 10.1KK254 pKa = 9.2KK255 pKa = 8.77TVAKK259 pKa = 10.17KK260 pKa = 9.94AAPKK264 pKa = 9.94KK265 pKa = 10.07AAAKK269 pKa = 9.86KK270 pKa = 9.67AAPKK274 pKa = 9.99KK275 pKa = 9.89AAPKK279 pKa = 9.63KK280 pKa = 9.25AAPKK284 pKa = 9.99KK285 pKa = 10.0VEE287 pKa = 4.22TKK289 pKa = 10.18KK290 pKa = 10.91VEE292 pKa = 4.31PKK294 pKa = 9.78TVAPTPKK301 pKa = 9.53PVASPAPRR309 pKa = 11.84PPVAQPAQRR318 pKa = 11.84PPLAQPSPSPARR330 pKa = 11.84PSSPGAGGTPPRR342 pKa = 11.84HH343 pKa = 5.71VSPGSAIPGYY353 pKa = 9.48QPPRR357 pKa = 11.84PPMPGSTPGPTPTPMPYY374 pKa = 9.6RR375 pKa = 11.84PSWARR380 pKa = 11.84DD381 pKa = 3.24DD382 pKa = 4.42DD383 pKa = 4.0
Molecular weight: 40.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
996689 |
37 |
3557 |
298.3 |
32.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.027 ± 0.064 | 0.874 ± 0.015 |
6.077 ± 0.045 | 6.214 ± 0.044 |
3.724 ± 0.031 | 8.491 ± 0.053 |
1.964 ± 0.021 | 4.944 ± 0.033 |
3.811 ± 0.037 | 9.882 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.335 ± 0.023 | 2.68 ± 0.029 |
5.286 ± 0.031 | 3.065 ± 0.024 |
6.612 ± 0.044 | 5.372 ± 0.03 |
5.421 ± 0.036 | 7.472 ± 0.041 |
1.339 ± 0.02 | 2.41 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
