
Flavobacteria bacterium (strain MS024-2A)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; unclassified Flavobacteriia
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
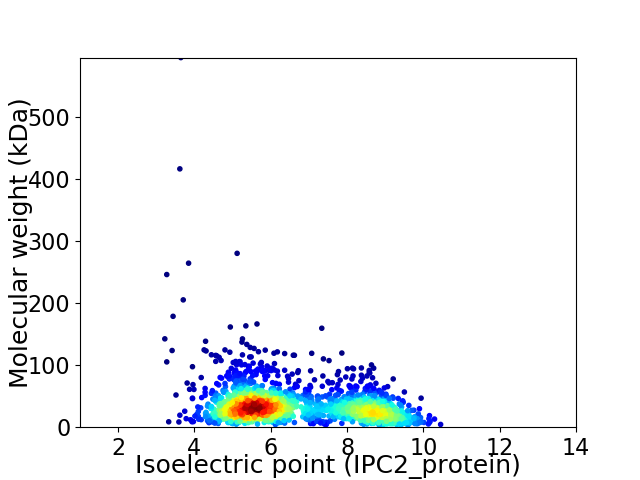
Virtual 2D-PAGE plot for 1770 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
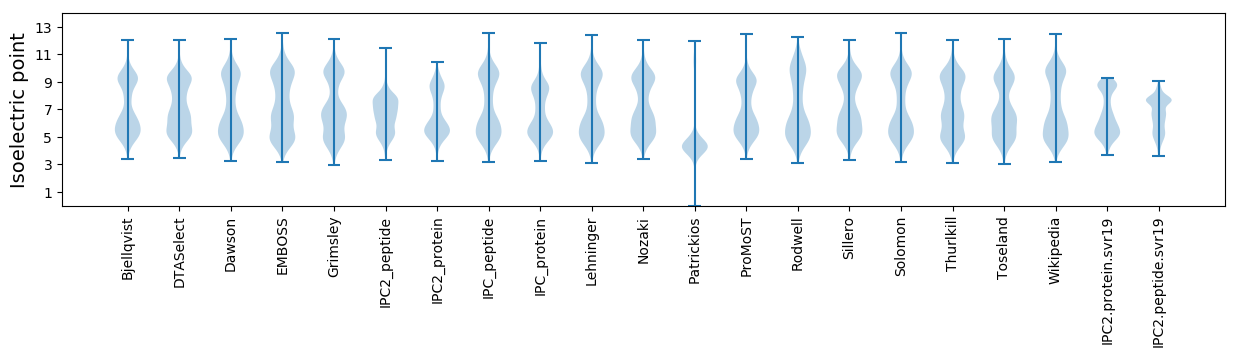
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0BIU0|C0BIU0_FLABM PSP1 domain protein OS=Flavobacteria bacterium (strain MS024-2A) OX=487796 GN=Flav2ADRAFT_0752 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.37RR3 pKa = 11.84ILYY6 pKa = 9.56LFILLGFNLVYY17 pKa = 10.63AQTEE21 pKa = 4.37TFVISPGGQVSNTVIDD37 pKa = 4.5NIGQTVTTSALSGGLTLSEE56 pKa = 3.91GFLYY60 pKa = 10.93NFDD63 pKa = 3.65FTTPTVILTDD73 pKa = 3.44TDD75 pKa = 4.24SNNLVSNSDD84 pKa = 3.51VVTITATFSEE94 pKa = 4.86SMAATPTLSLSGITTGALMTASASSSIWTYY124 pKa = 7.88TWTVSTTVTSTTATVSGTDD143 pKa = 3.46LSGNAYY149 pKa = 10.31AGTDD153 pKa = 3.34SLTFSIDD160 pKa = 3.34NLGPTVVLSNNSLNNRR176 pKa = 11.84VSNTQSVTLYY186 pKa = 9.6ATFSEE191 pKa = 4.71EE192 pKa = 4.93LDD194 pKa = 3.3ITPTISLSGITTDD207 pKa = 4.0AAMSNGNTIVLKK219 pKa = 10.51PNNGWTSSSGSFQDD233 pKa = 3.97FNGSSGSHH241 pKa = 7.06PYY243 pKa = 10.01LAEE246 pKa = 4.27DD247 pKa = 3.87LVIFSYY253 pKa = 11.21LDD255 pKa = 3.1NASIYY260 pKa = 11.01KK261 pKa = 10.32NFTISSGTVSVVVKK275 pKa = 10.58LDD277 pKa = 3.48YY278 pKa = 10.63KK279 pKa = 10.78KK280 pKa = 10.86SYY282 pKa = 11.1SEE284 pKa = 3.94DD285 pKa = 3.21TGQVKK290 pKa = 10.41LYY292 pKa = 10.54YY293 pKa = 10.09YY294 pKa = 10.31DD295 pKa = 4.39ANSTLLSTDD304 pKa = 2.86ASAVLTGTSDD314 pKa = 3.6FQNLNFQSSVPSGAVVVRR332 pKa = 11.84LEE334 pKa = 4.6LIQINEE340 pKa = 4.15SEE342 pKa = 4.5FWAGNYY348 pKa = 9.23GIRR351 pKa = 11.84YY352 pKa = 8.77RR353 pKa = 11.84DD354 pKa = 3.61LEE356 pKa = 4.03ITGVDD361 pKa = 3.13EE362 pKa = 4.18GTVYY366 pKa = 10.89GGNDD370 pKa = 2.74TWSYY374 pKa = 8.14TWTVSTTATSTTATVSATDD393 pKa = 3.49NAGNSYY399 pKa = 9.43TGSEE403 pKa = 4.13SMTFVIDD410 pKa = 3.32NEE412 pKa = 4.04RR413 pKa = 11.84PYY415 pKa = 11.31AVSTTLSNNLKK426 pKa = 10.32AGYY429 pKa = 9.85ISATDD434 pKa = 3.71TNTLTITFNEE444 pKa = 4.68EE445 pKa = 3.32INASTFTASDD455 pKa = 3.53VTILPSGYY463 pKa = 10.11FSISEE468 pKa = 4.28HH469 pKa = 5.8TSSDD473 pKa = 2.92NTIFIGYY480 pKa = 9.86LNVLSNYY487 pKa = 10.07AGVVTLTLNEE497 pKa = 3.97NSLEE501 pKa = 4.17DD502 pKa = 3.65FAGNANTVASISFISDD518 pKa = 3.18YY519 pKa = 10.57TVPTVTLTDD528 pKa = 3.5TDD530 pKa = 3.56SDD532 pKa = 4.01NLVSNSEE539 pKa = 4.34VVTLTATFSEE549 pKa = 4.86SMSATPTLSLTGIVSDD565 pKa = 5.03AEE567 pKa = 4.3MTATASDD574 pKa = 4.58SIWTYY579 pKa = 7.58TWTVSGSTVTSTTATVSGTDD599 pKa = 3.54LAGNAYY605 pKa = 10.37SGTDD609 pKa = 3.42SITFTIDD616 pKa = 2.8NSNPTLVSFTDD627 pKa = 3.51SDD629 pKa = 3.58TDD631 pKa = 3.7NYY633 pKa = 9.99VNDD636 pKa = 3.91YY637 pKa = 11.04TNVTLSATFSEE648 pKa = 5.1SMASSPTIKK657 pKa = 9.5IDD659 pKa = 2.98NGTNVILVNSASMTVSSSTSSATWTYY685 pKa = 10.36FWDD688 pKa = 3.92VPGGSDD694 pKa = 3.11GTYY697 pKa = 9.37FATVSGTDD705 pKa = 3.46LSGNAYY711 pKa = 9.23TGTEE715 pKa = 4.22SITFVVDD722 pKa = 3.11NTAPLIQSVTVTNTNSKK739 pKa = 10.3VLLTYY744 pKa = 10.6NEE746 pKa = 4.23PVQLYY751 pKa = 9.66DD752 pKa = 3.43PSYY755 pKa = 8.8FASNFSVTKK764 pKa = 9.57TGGTATVSYY773 pKa = 9.41TGYY776 pKa = 10.6SFSTTDD782 pKa = 3.18SNTVILNIIVTGEE795 pKa = 3.84PTGDD799 pKa = 3.34EE800 pKa = 4.18LLEE803 pKa = 4.15VGPVGASTLIDD814 pKa = 3.33KK815 pKa = 10.6AGNYY819 pKa = 10.05AMDD822 pKa = 4.1YY823 pKa = 10.63SDD825 pKa = 5.89SNQTSNTVYY834 pKa = 9.75LTNTPPKK841 pKa = 9.67FASTSVSSNNTSITIAFSEE860 pKa = 4.93GVTADD865 pKa = 3.3TSGTEE870 pKa = 3.86LSKK873 pKa = 10.06TDD875 pKa = 3.43FTLAVSGGAAVLASATPIALSKK897 pKa = 9.63TDD899 pKa = 2.96SKK901 pKa = 10.27TYY903 pKa = 11.17VLTTSYY909 pKa = 10.93SVPSNGSEE917 pKa = 3.75ILTVTPISSAVFDD930 pKa = 4.41SKK932 pKa = 10.71GTQINLSSAQSNTVQLNDD950 pKa = 3.27QAGPFITGATLDD962 pKa = 3.67SQNRR966 pKa = 11.84YY967 pKa = 8.89VDD969 pKa = 4.04FTFSEE974 pKa = 4.79GVYY977 pKa = 9.21ATASPTSAVTSSSFTLSQSSGPSYY1001 pKa = 11.42GMTISSITTAGGGVLSGGEE1020 pKa = 3.81TTLRR1024 pKa = 11.84FNLEE1028 pKa = 4.13SAVKK1032 pKa = 8.72PTGQEE1037 pKa = 3.61VFAITATDD1045 pKa = 3.31SSSVVDD1051 pKa = 3.71ASGNSMTVSQTNNTFQLKK1069 pKa = 9.4PPTSGGVSPKK1079 pKa = 10.37KK1080 pKa = 9.62STISAAPAQMIGNGINTAVITVQAKK1105 pKa = 10.57DD1106 pKa = 3.38SLGQNFLEE1114 pKa = 4.84GGYY1117 pKa = 10.28QITLFGPNGDD1127 pKa = 3.92LTTTDD1132 pKa = 3.86NQNGTYY1138 pKa = 9.46SASYY1142 pKa = 8.54TPEE1145 pKa = 5.94AITQDD1150 pKa = 3.69FQEE1153 pKa = 4.29LTFGFRR1159 pKa = 11.84VADD1162 pKa = 3.83TNGTPTALLQLYY1174 pKa = 10.07LDD1176 pKa = 4.1EE1177 pKa = 7.41DD1178 pKa = 4.32GDD1180 pKa = 4.25GVYY1183 pKa = 10.69NINDD1187 pKa = 3.5ACLGTEE1193 pKa = 4.17AGLEE1197 pKa = 4.02VDD1199 pKa = 4.02ATGCALNQLDD1209 pKa = 4.3SDD1211 pKa = 4.09NDD1213 pKa = 4.07GVFDD1217 pKa = 6.35DD1218 pKa = 6.17LDD1220 pKa = 4.03EE1221 pKa = 5.91CPDD1224 pKa = 3.64TPEE1227 pKa = 3.94QEE1229 pKa = 4.33LNNVQGTPTYY1239 pKa = 10.93GEE1241 pKa = 5.17LIPTVVDD1248 pKa = 3.79EE1249 pKa = 4.9KK1250 pKa = 11.53GCGASQRR1257 pKa = 11.84DD1258 pKa = 3.69TDD1260 pKa = 3.45SDD1262 pKa = 4.72GIVDD1266 pKa = 4.05TEE1268 pKa = 4.71DD1269 pKa = 3.33NCIEE1273 pKa = 4.25TANADD1278 pKa = 3.69QADD1281 pKa = 3.93TDD1283 pKa = 3.96GDD1285 pKa = 4.53GIGDD1289 pKa = 3.67VCDD1292 pKa = 3.77TDD1294 pKa = 4.05NPLPEE1299 pKa = 4.07ITTTEE1304 pKa = 3.78IKK1306 pKa = 10.3FVQLPQNGTTVGKK1319 pKa = 10.34IEE1321 pKa = 4.12ATDD1324 pKa = 3.8PDD1326 pKa = 4.88GEE1328 pKa = 4.6TLTFTQSGDD1337 pKa = 3.08NFTGVLSIASDD1348 pKa = 3.71GSITVSSGALLTFDD1362 pKa = 3.47STYY1365 pKa = 11.28NGASLGFTVSDD1376 pKa = 3.93GKK1378 pKa = 10.76NDD1380 pKa = 3.21VHH1382 pKa = 8.03GSVKK1386 pKa = 10.24ILIEE1390 pKa = 4.22DD1391 pKa = 3.55VPRR1394 pKa = 11.84PPEE1397 pKa = 3.51ISIITLEE1404 pKa = 3.81ISEE1407 pKa = 4.33DD1408 pKa = 3.95AEE1410 pKa = 4.14VGTIVGFVEE1419 pKa = 5.28AKK1421 pKa = 10.67DD1422 pKa = 3.62PMGGQIVSISLQGDD1436 pKa = 3.87GFIEE1440 pKa = 4.59LKK1442 pKa = 10.97DD1443 pKa = 3.82GVLKK1447 pKa = 8.13TTQEE1451 pKa = 3.63LDD1453 pKa = 3.47YY1454 pKa = 10.88EE1455 pKa = 4.66EE1456 pKa = 4.66NTIHH1460 pKa = 7.41PFTITAQASDD1470 pKa = 3.75RR1471 pKa = 11.84SDD1473 pKa = 3.72GPGLTGSKK1481 pKa = 10.13SEE1483 pKa = 4.18SLRR1486 pKa = 11.84VSDD1489 pKa = 5.16IPNATYY1495 pKa = 9.69TGRR1498 pKa = 11.84FFISIFNVEE1507 pKa = 4.71DD1508 pKa = 3.28EE1509 pKa = 4.46TLGAKK1514 pKa = 9.38VDD1516 pKa = 3.17HH1517 pKa = 6.38RR1518 pKa = 11.84RR1519 pKa = 11.84YY1520 pKa = 9.97FNPYY1524 pKa = 8.43NKK1526 pKa = 9.82NVGKK1530 pKa = 9.69WKK1532 pKa = 9.64IKK1534 pKa = 10.18KK1535 pKa = 9.34RR1536 pKa = 11.84VAGGADD1542 pKa = 3.28ADD1544 pKa = 3.84KK1545 pKa = 9.91FTIKK1549 pKa = 9.5TRR1551 pKa = 11.84SKK1553 pKa = 9.78TEE1555 pKa = 3.69QKK1557 pKa = 10.74NDD1559 pKa = 3.98DD1560 pKa = 4.6PIEE1563 pKa = 4.62DD1564 pKa = 3.95EE1565 pKa = 4.56NEE1567 pKa = 4.13DD1568 pKa = 3.73YY1569 pKa = 11.43LDD1571 pKa = 5.53FITPPDD1577 pKa = 3.73YY1578 pKa = 11.24EE1579 pKa = 4.46NPGDD1583 pKa = 3.97ANGDD1587 pKa = 3.53NIYY1590 pKa = 10.04EE1591 pKa = 4.23VEE1593 pKa = 4.09VEE1595 pKa = 4.49YY1596 pKa = 11.62VNTDD1600 pKa = 3.06DD1601 pKa = 5.26GAPEE1605 pKa = 3.94VPIVVTQTNIQVPEE1619 pKa = 4.36GSTTTIEE1626 pKa = 4.29LQSQPVLPTDD1636 pKa = 4.88DD1637 pKa = 4.22NDD1639 pKa = 4.08GDD1641 pKa = 4.34GVPDD1645 pKa = 5.11IIDD1648 pKa = 3.84NSPLVANADD1657 pKa = 3.62QSDD1660 pKa = 3.7EE1661 pKa = 4.92DD1662 pKa = 4.55GDD1664 pKa = 4.48GVGDD1668 pKa = 3.68VSDD1671 pKa = 5.72DD1672 pKa = 3.75FDD1674 pKa = 6.61HH1675 pKa = 7.6DD1676 pKa = 4.74GVWNPFDD1683 pKa = 3.8TCPDD1687 pKa = 3.35TSLGEE1692 pKa = 4.1IVDD1695 pKa = 4.2SNGCLIYY1702 pKa = 9.64YY1703 pKa = 9.69IPSSNFSISKK1713 pKa = 8.28TEE1715 pKa = 3.66KK1716 pKa = 10.19CAGTNSINIEE1726 pKa = 4.16VVDD1729 pKa = 3.66TSITYY1734 pKa = 9.4NVSVSGAATLSDD1746 pKa = 4.05SFSSSSWSLDD1756 pKa = 3.2NLSAGVYY1763 pKa = 8.61SICITVEE1770 pKa = 4.28GVSSAEE1776 pKa = 3.78FEE1778 pKa = 4.28RR1779 pKa = 11.84CFEE1782 pKa = 4.24VTISEE1787 pKa = 4.44PDD1789 pKa = 3.67PLIVNSMFNKK1799 pKa = 9.88NDD1801 pKa = 3.19QTITFGLNGGVSYY1814 pKa = 10.65DD1815 pKa = 3.2ITHH1818 pKa = 6.75NGITTQTTKK1827 pKa = 10.74SSYY1830 pKa = 9.45TVSLAKK1836 pKa = 10.16GLNRR1840 pKa = 11.84ISISTGIEE1848 pKa = 3.95CQGLFEE1854 pKa = 4.1NTYY1857 pKa = 10.03LNSYY1861 pKa = 6.82EE1862 pKa = 4.25VKK1864 pKa = 10.17YY1865 pKa = 10.88APNPFSSEE1873 pKa = 3.81LTFTIGGEE1881 pKa = 3.88DD1882 pKa = 2.92RR1883 pKa = 11.84VFGLEE1888 pKa = 3.96VYY1890 pKa = 9.37TPSGQLIDD1898 pKa = 3.72QKK1900 pKa = 11.4VITLPFGIRR1909 pKa = 11.84YY1910 pKa = 7.34YY1911 pKa = 10.5TLQTDD1916 pKa = 3.63NYY1918 pKa = 8.57KK1919 pKa = 10.45QGGYY1923 pKa = 9.96FIRR1926 pKa = 11.84VKK1928 pKa = 10.78GSTIDD1933 pKa = 3.71QSFQVIKK1940 pKa = 10.3EE1941 pKa = 3.98
MM1 pKa = 7.48KK2 pKa = 10.37RR3 pKa = 11.84ILYY6 pKa = 9.56LFILLGFNLVYY17 pKa = 10.63AQTEE21 pKa = 4.37TFVISPGGQVSNTVIDD37 pKa = 4.5NIGQTVTTSALSGGLTLSEE56 pKa = 3.91GFLYY60 pKa = 10.93NFDD63 pKa = 3.65FTTPTVILTDD73 pKa = 3.44TDD75 pKa = 4.24SNNLVSNSDD84 pKa = 3.51VVTITATFSEE94 pKa = 4.86SMAATPTLSLSGITTGALMTASASSSIWTYY124 pKa = 7.88TWTVSTTVTSTTATVSGTDD143 pKa = 3.46LSGNAYY149 pKa = 10.31AGTDD153 pKa = 3.34SLTFSIDD160 pKa = 3.34NLGPTVVLSNNSLNNRR176 pKa = 11.84VSNTQSVTLYY186 pKa = 9.6ATFSEE191 pKa = 4.71EE192 pKa = 4.93LDD194 pKa = 3.3ITPTISLSGITTDD207 pKa = 4.0AAMSNGNTIVLKK219 pKa = 10.51PNNGWTSSSGSFQDD233 pKa = 3.97FNGSSGSHH241 pKa = 7.06PYY243 pKa = 10.01LAEE246 pKa = 4.27DD247 pKa = 3.87LVIFSYY253 pKa = 11.21LDD255 pKa = 3.1NASIYY260 pKa = 11.01KK261 pKa = 10.32NFTISSGTVSVVVKK275 pKa = 10.58LDD277 pKa = 3.48YY278 pKa = 10.63KK279 pKa = 10.78KK280 pKa = 10.86SYY282 pKa = 11.1SEE284 pKa = 3.94DD285 pKa = 3.21TGQVKK290 pKa = 10.41LYY292 pKa = 10.54YY293 pKa = 10.09YY294 pKa = 10.31DD295 pKa = 4.39ANSTLLSTDD304 pKa = 2.86ASAVLTGTSDD314 pKa = 3.6FQNLNFQSSVPSGAVVVRR332 pKa = 11.84LEE334 pKa = 4.6LIQINEE340 pKa = 4.15SEE342 pKa = 4.5FWAGNYY348 pKa = 9.23GIRR351 pKa = 11.84YY352 pKa = 8.77RR353 pKa = 11.84DD354 pKa = 3.61LEE356 pKa = 4.03ITGVDD361 pKa = 3.13EE362 pKa = 4.18GTVYY366 pKa = 10.89GGNDD370 pKa = 2.74TWSYY374 pKa = 8.14TWTVSTTATSTTATVSATDD393 pKa = 3.49NAGNSYY399 pKa = 9.43TGSEE403 pKa = 4.13SMTFVIDD410 pKa = 3.32NEE412 pKa = 4.04RR413 pKa = 11.84PYY415 pKa = 11.31AVSTTLSNNLKK426 pKa = 10.32AGYY429 pKa = 9.85ISATDD434 pKa = 3.71TNTLTITFNEE444 pKa = 4.68EE445 pKa = 3.32INASTFTASDD455 pKa = 3.53VTILPSGYY463 pKa = 10.11FSISEE468 pKa = 4.28HH469 pKa = 5.8TSSDD473 pKa = 2.92NTIFIGYY480 pKa = 9.86LNVLSNYY487 pKa = 10.07AGVVTLTLNEE497 pKa = 3.97NSLEE501 pKa = 4.17DD502 pKa = 3.65FAGNANTVASISFISDD518 pKa = 3.18YY519 pKa = 10.57TVPTVTLTDD528 pKa = 3.5TDD530 pKa = 3.56SDD532 pKa = 4.01NLVSNSEE539 pKa = 4.34VVTLTATFSEE549 pKa = 4.86SMSATPTLSLTGIVSDD565 pKa = 5.03AEE567 pKa = 4.3MTATASDD574 pKa = 4.58SIWTYY579 pKa = 7.58TWTVSGSTVTSTTATVSGTDD599 pKa = 3.54LAGNAYY605 pKa = 10.37SGTDD609 pKa = 3.42SITFTIDD616 pKa = 2.8NSNPTLVSFTDD627 pKa = 3.51SDD629 pKa = 3.58TDD631 pKa = 3.7NYY633 pKa = 9.99VNDD636 pKa = 3.91YY637 pKa = 11.04TNVTLSATFSEE648 pKa = 5.1SMASSPTIKK657 pKa = 9.5IDD659 pKa = 2.98NGTNVILVNSASMTVSSSTSSATWTYY685 pKa = 10.36FWDD688 pKa = 3.92VPGGSDD694 pKa = 3.11GTYY697 pKa = 9.37FATVSGTDD705 pKa = 3.46LSGNAYY711 pKa = 9.23TGTEE715 pKa = 4.22SITFVVDD722 pKa = 3.11NTAPLIQSVTVTNTNSKK739 pKa = 10.3VLLTYY744 pKa = 10.6NEE746 pKa = 4.23PVQLYY751 pKa = 9.66DD752 pKa = 3.43PSYY755 pKa = 8.8FASNFSVTKK764 pKa = 9.57TGGTATVSYY773 pKa = 9.41TGYY776 pKa = 10.6SFSTTDD782 pKa = 3.18SNTVILNIIVTGEE795 pKa = 3.84PTGDD799 pKa = 3.34EE800 pKa = 4.18LLEE803 pKa = 4.15VGPVGASTLIDD814 pKa = 3.33KK815 pKa = 10.6AGNYY819 pKa = 10.05AMDD822 pKa = 4.1YY823 pKa = 10.63SDD825 pKa = 5.89SNQTSNTVYY834 pKa = 9.75LTNTPPKK841 pKa = 9.67FASTSVSSNNTSITIAFSEE860 pKa = 4.93GVTADD865 pKa = 3.3TSGTEE870 pKa = 3.86LSKK873 pKa = 10.06TDD875 pKa = 3.43FTLAVSGGAAVLASATPIALSKK897 pKa = 9.63TDD899 pKa = 2.96SKK901 pKa = 10.27TYY903 pKa = 11.17VLTTSYY909 pKa = 10.93SVPSNGSEE917 pKa = 3.75ILTVTPISSAVFDD930 pKa = 4.41SKK932 pKa = 10.71GTQINLSSAQSNTVQLNDD950 pKa = 3.27QAGPFITGATLDD962 pKa = 3.67SQNRR966 pKa = 11.84YY967 pKa = 8.89VDD969 pKa = 4.04FTFSEE974 pKa = 4.79GVYY977 pKa = 9.21ATASPTSAVTSSSFTLSQSSGPSYY1001 pKa = 11.42GMTISSITTAGGGVLSGGEE1020 pKa = 3.81TTLRR1024 pKa = 11.84FNLEE1028 pKa = 4.13SAVKK1032 pKa = 8.72PTGQEE1037 pKa = 3.61VFAITATDD1045 pKa = 3.31SSSVVDD1051 pKa = 3.71ASGNSMTVSQTNNTFQLKK1069 pKa = 9.4PPTSGGVSPKK1079 pKa = 10.37KK1080 pKa = 9.62STISAAPAQMIGNGINTAVITVQAKK1105 pKa = 10.57DD1106 pKa = 3.38SLGQNFLEE1114 pKa = 4.84GGYY1117 pKa = 10.28QITLFGPNGDD1127 pKa = 3.92LTTTDD1132 pKa = 3.86NQNGTYY1138 pKa = 9.46SASYY1142 pKa = 8.54TPEE1145 pKa = 5.94AITQDD1150 pKa = 3.69FQEE1153 pKa = 4.29LTFGFRR1159 pKa = 11.84VADD1162 pKa = 3.83TNGTPTALLQLYY1174 pKa = 10.07LDD1176 pKa = 4.1EE1177 pKa = 7.41DD1178 pKa = 4.32GDD1180 pKa = 4.25GVYY1183 pKa = 10.69NINDD1187 pKa = 3.5ACLGTEE1193 pKa = 4.17AGLEE1197 pKa = 4.02VDD1199 pKa = 4.02ATGCALNQLDD1209 pKa = 4.3SDD1211 pKa = 4.09NDD1213 pKa = 4.07GVFDD1217 pKa = 6.35DD1218 pKa = 6.17LDD1220 pKa = 4.03EE1221 pKa = 5.91CPDD1224 pKa = 3.64TPEE1227 pKa = 3.94QEE1229 pKa = 4.33LNNVQGTPTYY1239 pKa = 10.93GEE1241 pKa = 5.17LIPTVVDD1248 pKa = 3.79EE1249 pKa = 4.9KK1250 pKa = 11.53GCGASQRR1257 pKa = 11.84DD1258 pKa = 3.69TDD1260 pKa = 3.45SDD1262 pKa = 4.72GIVDD1266 pKa = 4.05TEE1268 pKa = 4.71DD1269 pKa = 3.33NCIEE1273 pKa = 4.25TANADD1278 pKa = 3.69QADD1281 pKa = 3.93TDD1283 pKa = 3.96GDD1285 pKa = 4.53GIGDD1289 pKa = 3.67VCDD1292 pKa = 3.77TDD1294 pKa = 4.05NPLPEE1299 pKa = 4.07ITTTEE1304 pKa = 3.78IKK1306 pKa = 10.3FVQLPQNGTTVGKK1319 pKa = 10.34IEE1321 pKa = 4.12ATDD1324 pKa = 3.8PDD1326 pKa = 4.88GEE1328 pKa = 4.6TLTFTQSGDD1337 pKa = 3.08NFTGVLSIASDD1348 pKa = 3.71GSITVSSGALLTFDD1362 pKa = 3.47STYY1365 pKa = 11.28NGASLGFTVSDD1376 pKa = 3.93GKK1378 pKa = 10.76NDD1380 pKa = 3.21VHH1382 pKa = 8.03GSVKK1386 pKa = 10.24ILIEE1390 pKa = 4.22DD1391 pKa = 3.55VPRR1394 pKa = 11.84PPEE1397 pKa = 3.51ISIITLEE1404 pKa = 3.81ISEE1407 pKa = 4.33DD1408 pKa = 3.95AEE1410 pKa = 4.14VGTIVGFVEE1419 pKa = 5.28AKK1421 pKa = 10.67DD1422 pKa = 3.62PMGGQIVSISLQGDD1436 pKa = 3.87GFIEE1440 pKa = 4.59LKK1442 pKa = 10.97DD1443 pKa = 3.82GVLKK1447 pKa = 8.13TTQEE1451 pKa = 3.63LDD1453 pKa = 3.47YY1454 pKa = 10.88EE1455 pKa = 4.66EE1456 pKa = 4.66NTIHH1460 pKa = 7.41PFTITAQASDD1470 pKa = 3.75RR1471 pKa = 11.84SDD1473 pKa = 3.72GPGLTGSKK1481 pKa = 10.13SEE1483 pKa = 4.18SLRR1486 pKa = 11.84VSDD1489 pKa = 5.16IPNATYY1495 pKa = 9.69TGRR1498 pKa = 11.84FFISIFNVEE1507 pKa = 4.71DD1508 pKa = 3.28EE1509 pKa = 4.46TLGAKK1514 pKa = 9.38VDD1516 pKa = 3.17HH1517 pKa = 6.38RR1518 pKa = 11.84RR1519 pKa = 11.84YY1520 pKa = 9.97FNPYY1524 pKa = 8.43NKK1526 pKa = 9.82NVGKK1530 pKa = 9.69WKK1532 pKa = 9.64IKK1534 pKa = 10.18KK1535 pKa = 9.34RR1536 pKa = 11.84VAGGADD1542 pKa = 3.28ADD1544 pKa = 3.84KK1545 pKa = 9.91FTIKK1549 pKa = 9.5TRR1551 pKa = 11.84SKK1553 pKa = 9.78TEE1555 pKa = 3.69QKK1557 pKa = 10.74NDD1559 pKa = 3.98DD1560 pKa = 4.6PIEE1563 pKa = 4.62DD1564 pKa = 3.95EE1565 pKa = 4.56NEE1567 pKa = 4.13DD1568 pKa = 3.73YY1569 pKa = 11.43LDD1571 pKa = 5.53FITPPDD1577 pKa = 3.73YY1578 pKa = 11.24EE1579 pKa = 4.46NPGDD1583 pKa = 3.97ANGDD1587 pKa = 3.53NIYY1590 pKa = 10.04EE1591 pKa = 4.23VEE1593 pKa = 4.09VEE1595 pKa = 4.49YY1596 pKa = 11.62VNTDD1600 pKa = 3.06DD1601 pKa = 5.26GAPEE1605 pKa = 3.94VPIVVTQTNIQVPEE1619 pKa = 4.36GSTTTIEE1626 pKa = 4.29LQSQPVLPTDD1636 pKa = 4.88DD1637 pKa = 4.22NDD1639 pKa = 4.08GDD1641 pKa = 4.34GVPDD1645 pKa = 5.11IIDD1648 pKa = 3.84NSPLVANADD1657 pKa = 3.62QSDD1660 pKa = 3.7EE1661 pKa = 4.92DD1662 pKa = 4.55GDD1664 pKa = 4.48GVGDD1668 pKa = 3.68VSDD1671 pKa = 5.72DD1672 pKa = 3.75FDD1674 pKa = 6.61HH1675 pKa = 7.6DD1676 pKa = 4.74GVWNPFDD1683 pKa = 3.8TCPDD1687 pKa = 3.35TSLGEE1692 pKa = 4.1IVDD1695 pKa = 4.2SNGCLIYY1702 pKa = 9.64YY1703 pKa = 9.69IPSSNFSISKK1713 pKa = 8.28TEE1715 pKa = 3.66KK1716 pKa = 10.19CAGTNSINIEE1726 pKa = 4.16VVDD1729 pKa = 3.66TSITYY1734 pKa = 9.4NVSVSGAATLSDD1746 pKa = 4.05SFSSSSWSLDD1756 pKa = 3.2NLSAGVYY1763 pKa = 8.61SICITVEE1770 pKa = 4.28GVSSAEE1776 pKa = 3.78FEE1778 pKa = 4.28RR1779 pKa = 11.84CFEE1782 pKa = 4.24VTISEE1787 pKa = 4.44PDD1789 pKa = 3.67PLIVNSMFNKK1799 pKa = 9.88NDD1801 pKa = 3.19QTITFGLNGGVSYY1814 pKa = 10.65DD1815 pKa = 3.2ITHH1818 pKa = 6.75NGITTQTTKK1827 pKa = 10.74SSYY1830 pKa = 9.45TVSLAKK1836 pKa = 10.16GLNRR1840 pKa = 11.84ISISTGIEE1848 pKa = 3.95CQGLFEE1854 pKa = 4.1NTYY1857 pKa = 10.03LNSYY1861 pKa = 6.82EE1862 pKa = 4.25VKK1864 pKa = 10.17YY1865 pKa = 10.88APNPFSSEE1873 pKa = 3.81LTFTIGGEE1881 pKa = 3.88DD1882 pKa = 2.92RR1883 pKa = 11.84VFGLEE1888 pKa = 3.96VYY1890 pKa = 9.37TPSGQLIDD1898 pKa = 3.72QKK1900 pKa = 11.4VITLPFGIRR1909 pKa = 11.84YY1910 pKa = 7.34YY1911 pKa = 10.5TLQTDD1916 pKa = 3.63NYY1918 pKa = 8.57KK1919 pKa = 10.45QGGYY1923 pKa = 9.96FIRR1926 pKa = 11.84VKK1928 pKa = 10.78GSTIDD1933 pKa = 3.71QSFQVIKK1940 pKa = 10.3EE1941 pKa = 3.98
Molecular weight: 205.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0BKI2|C0BKI2_FLABM Histidine triad (HIT) protein OS=Flavobacteria bacterium (strain MS024-2A) OX=487796 GN=Flav2ADRAFT_0055 PE=4 SV=1
MM1 pKa = 7.02VVVAHH6 pKa = 6.61LVRR9 pKa = 11.84APDD12 pKa = 3.98FGTGDD17 pKa = 4.72LISSRR22 pKa = 11.84VLKK25 pKa = 10.95NSFPFNRR32 pKa = 11.84FTNIFSS38 pKa = 3.58
MM1 pKa = 7.02VVVAHH6 pKa = 6.61LVRR9 pKa = 11.84APDD12 pKa = 3.98FGTGDD17 pKa = 4.72LISSRR22 pKa = 11.84VLKK25 pKa = 10.95NSFPFNRR32 pKa = 11.84FTNIFSS38 pKa = 3.58
Molecular weight: 4.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
592508 |
32 |
5671 |
334.8 |
37.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.538 ± 0.064 | 0.744 ± 0.021 |
5.35 ± 0.095 | 6.492 ± 0.063 |
5.287 ± 0.051 | 6.78 ± 0.057 |
1.874 ± 0.034 | 7.842 ± 0.049 |
7.149 ± 0.095 | 9.762 ± 0.082 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.19 ± 0.037 | 5.574 ± 0.063 |
3.76 ± 0.031 | 3.697 ± 0.041 |
3.559 ± 0.046 | 6.968 ± 0.063 |
5.601 ± 0.112 | 5.998 ± 0.047 |
1.159 ± 0.023 | 3.678 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
