
Algibacter lectus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group;
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
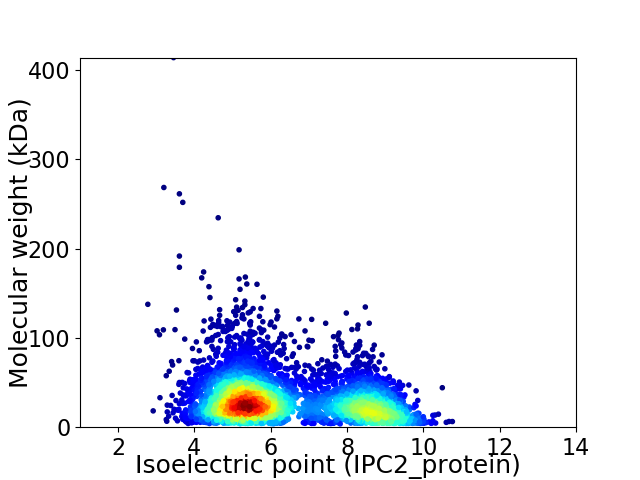
Virtual 2D-PAGE plot for 4609 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A090VNL6|A0A090VNL6_9FLAO Uncharacterized protein OS=Algibacter lectus OX=221126 GN=JCM19300_8 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.43FKK4 pKa = 10.97LFNPLYY10 pKa = 10.57LFTLTLFVASCSSDD24 pKa = 3.79DD25 pKa = 4.48DD26 pKa = 5.15GGDD29 pKa = 3.19TSEE32 pKa = 5.1SIEE35 pKa = 3.93KK36 pKa = 10.51SAVIEE41 pKa = 4.17NYY43 pKa = 10.98ANLVYY48 pKa = 10.42QSYY51 pKa = 10.75LDD53 pKa = 4.03SYY55 pKa = 9.24TGAVSLQTAVDD66 pKa = 3.8VFIEE70 pKa = 4.52TPTDD74 pKa = 3.48ANFTAVKK81 pKa = 8.58TAWLSARR88 pKa = 11.84DD89 pKa = 3.55IYY91 pKa = 11.0GQTEE95 pKa = 4.59AYY97 pKa = 9.73RR98 pKa = 11.84EE99 pKa = 4.25CNGPIDD105 pKa = 4.23TEE107 pKa = 4.15NEE109 pKa = 3.55AWSVGNEE116 pKa = 4.05GQLNAWPIDD125 pKa = 3.58EE126 pKa = 5.38SYY128 pKa = 11.03IDD130 pKa = 3.85YY131 pKa = 11.01VAFGTEE137 pKa = 4.14AYY139 pKa = 9.8AGSFTSIISNTSVTIDD155 pKa = 3.47EE156 pKa = 4.56ATLASLNEE164 pKa = 3.94EE165 pKa = 4.35TTDD168 pKa = 3.92KK169 pKa = 11.12SISTGWHH176 pKa = 6.0AIEE179 pKa = 4.24FLLWGQDD186 pKa = 2.97NTLPSEE192 pKa = 4.41DD193 pKa = 4.38LPGQRR198 pKa = 11.84EE199 pKa = 4.15YY200 pKa = 11.29TDD202 pKa = 3.22YY203 pKa = 8.58TTADD207 pKa = 3.66DD208 pKa = 4.01ADD210 pKa = 3.71RR211 pKa = 11.84RR212 pKa = 11.84ADD214 pKa = 3.72YY215 pKa = 10.72LQAATDD221 pKa = 4.28LLVSDD226 pKa = 5.61LNALTTTWAPNGTYY240 pKa = 8.81RR241 pKa = 11.84TVFEE245 pKa = 5.1ALDD248 pKa = 3.51EE249 pKa = 4.46DD250 pKa = 4.25VALKK254 pKa = 10.47QFINGAFFIAGDD266 pKa = 3.86EE267 pKa = 4.2LSSEE271 pKa = 4.5RR272 pKa = 11.84IIAPVDD278 pKa = 3.49STDD281 pKa = 4.4GIDD284 pKa = 4.43GLGQEE289 pKa = 5.39DD290 pKa = 4.13EE291 pKa = 4.83HH292 pKa = 9.27SCFSDD297 pKa = 3.36NTHH300 pKa = 6.57KK301 pKa = 10.9DD302 pKa = 2.46IWANAQGVYY311 pKa = 10.54NVVFGAYY318 pKa = 9.96GNVSGDD324 pKa = 3.17SFYY327 pKa = 11.68DD328 pKa = 3.51LVKK331 pKa = 10.98EE332 pKa = 3.92KK333 pKa = 10.89DD334 pKa = 3.05AAQAEE339 pKa = 4.48KK340 pKa = 10.85LKK342 pKa = 10.99DD343 pKa = 3.5AADD346 pKa = 3.55EE347 pKa = 4.19AMEE350 pKa = 4.36KK351 pKa = 10.73VNAIANNSEE360 pKa = 3.88PFDD363 pKa = 3.86YY364 pKa = 11.12LITLEE369 pKa = 4.55NSTDD373 pKa = 3.39ANFGVVMQSVVALQDD388 pKa = 3.11WADD391 pKa = 4.41EE392 pKa = 4.27ISASATVIGINLL404 pKa = 3.48
MM1 pKa = 7.49KK2 pKa = 10.43FKK4 pKa = 10.97LFNPLYY10 pKa = 10.57LFTLTLFVASCSSDD24 pKa = 3.79DD25 pKa = 4.48DD26 pKa = 5.15GGDD29 pKa = 3.19TSEE32 pKa = 5.1SIEE35 pKa = 3.93KK36 pKa = 10.51SAVIEE41 pKa = 4.17NYY43 pKa = 10.98ANLVYY48 pKa = 10.42QSYY51 pKa = 10.75LDD53 pKa = 4.03SYY55 pKa = 9.24TGAVSLQTAVDD66 pKa = 3.8VFIEE70 pKa = 4.52TPTDD74 pKa = 3.48ANFTAVKK81 pKa = 8.58TAWLSARR88 pKa = 11.84DD89 pKa = 3.55IYY91 pKa = 11.0GQTEE95 pKa = 4.59AYY97 pKa = 9.73RR98 pKa = 11.84EE99 pKa = 4.25CNGPIDD105 pKa = 4.23TEE107 pKa = 4.15NEE109 pKa = 3.55AWSVGNEE116 pKa = 4.05GQLNAWPIDD125 pKa = 3.58EE126 pKa = 5.38SYY128 pKa = 11.03IDD130 pKa = 3.85YY131 pKa = 11.01VAFGTEE137 pKa = 4.14AYY139 pKa = 9.8AGSFTSIISNTSVTIDD155 pKa = 3.47EE156 pKa = 4.56ATLASLNEE164 pKa = 3.94EE165 pKa = 4.35TTDD168 pKa = 3.92KK169 pKa = 11.12SISTGWHH176 pKa = 6.0AIEE179 pKa = 4.24FLLWGQDD186 pKa = 2.97NTLPSEE192 pKa = 4.41DD193 pKa = 4.38LPGQRR198 pKa = 11.84EE199 pKa = 4.15YY200 pKa = 11.29TDD202 pKa = 3.22YY203 pKa = 8.58TTADD207 pKa = 3.66DD208 pKa = 4.01ADD210 pKa = 3.71RR211 pKa = 11.84RR212 pKa = 11.84ADD214 pKa = 3.72YY215 pKa = 10.72LQAATDD221 pKa = 4.28LLVSDD226 pKa = 5.61LNALTTTWAPNGTYY240 pKa = 8.81RR241 pKa = 11.84TVFEE245 pKa = 5.1ALDD248 pKa = 3.51EE249 pKa = 4.46DD250 pKa = 4.25VALKK254 pKa = 10.47QFINGAFFIAGDD266 pKa = 3.86EE267 pKa = 4.2LSSEE271 pKa = 4.5RR272 pKa = 11.84IIAPVDD278 pKa = 3.49STDD281 pKa = 4.4GIDD284 pKa = 4.43GLGQEE289 pKa = 5.39DD290 pKa = 4.13EE291 pKa = 4.83HH292 pKa = 9.27SCFSDD297 pKa = 3.36NTHH300 pKa = 6.57KK301 pKa = 10.9DD302 pKa = 2.46IWANAQGVYY311 pKa = 10.54NVVFGAYY318 pKa = 9.96GNVSGDD324 pKa = 3.17SFYY327 pKa = 11.68DD328 pKa = 3.51LVKK331 pKa = 10.98EE332 pKa = 3.92KK333 pKa = 10.89DD334 pKa = 3.05AAQAEE339 pKa = 4.48KK340 pKa = 10.85LKK342 pKa = 10.99DD343 pKa = 3.5AADD346 pKa = 3.55EE347 pKa = 4.19AMEE350 pKa = 4.36KK351 pKa = 10.73VNAIANNSEE360 pKa = 3.88PFDD363 pKa = 3.86YY364 pKa = 11.12LITLEE369 pKa = 4.55NSTDD373 pKa = 3.39ANFGVVMQSVVALQDD388 pKa = 3.11WADD391 pKa = 4.41EE392 pKa = 4.27ISASATVIGINLL404 pKa = 3.48
Molecular weight: 44.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A090WUR1|A0A090WUR1_9FLAO Glycosyltransferase OS=Algibacter lectus OX=221126 GN=JCM19274_1496 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
Molecular weight: 6.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1356275 |
37 |
3966 |
294.3 |
33.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.396 ± 0.035 | 0.742 ± 0.014 |
5.849 ± 0.033 | 6.487 ± 0.032 |
5.217 ± 0.029 | 6.34 ± 0.038 |
1.746 ± 0.019 | 7.803 ± 0.039 |
7.732 ± 0.052 | 9.128 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.112 ± 0.018 | 6.561 ± 0.042 |
3.303 ± 0.023 | 3.182 ± 0.02 |
3.169 ± 0.021 | 6.646 ± 0.033 |
6.097 ± 0.042 | 6.298 ± 0.031 |
1.089 ± 0.013 | 4.105 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
