
Pseudooceanicola nitratireducens
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pseudooceanicola
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
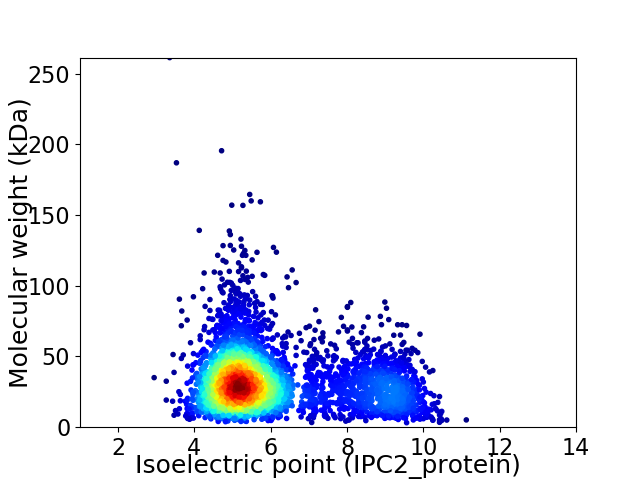
Virtual 2D-PAGE plot for 3828 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I1N3D5|A0A1I1N3D5_9RHOB Hydrogenase maturation protease OS=Pseudooceanicola nitratireducens OX=517719 GN=SAMN05421762_2745 PE=3 SV=1
MM1 pKa = 8.02ADD3 pKa = 3.23GKK5 pKa = 11.19YY6 pKa = 10.22DD7 pKa = 3.6GSSGSDD13 pKa = 3.09NINRR17 pKa = 11.84TSVDD21 pKa = 3.09GGGDD25 pKa = 3.42QVDD28 pKa = 4.62GPDD31 pKa = 3.74GLNDD35 pKa = 3.89SIYY38 pKa = 10.76GYY40 pKa = 10.56GGNDD44 pKa = 3.33TIEE47 pKa = 4.37AGRR50 pKa = 11.84GSDD53 pKa = 3.01TVYY56 pKa = 10.78GGEE59 pKa = 4.19GADD62 pKa = 3.76SIEE65 pKa = 4.16GRR67 pKa = 11.84SGDD70 pKa = 3.61DD71 pKa = 3.16VLYY74 pKa = 11.02GGAGRR79 pKa = 11.84DD80 pKa = 3.9TIEE83 pKa = 4.66GDD85 pKa = 3.19AGNDD89 pKa = 3.6TIYY92 pKa = 10.96VGSGDD97 pKa = 3.82DD98 pKa = 4.52DD99 pKa = 4.26IYY101 pKa = 11.64GEE103 pKa = 4.68SGDD106 pKa = 3.95DD107 pKa = 4.13LIRR110 pKa = 11.84VTRR113 pKa = 11.84GTSAWIDD120 pKa = 3.73GQSGDD125 pKa = 4.07DD126 pKa = 3.54TLDD129 pKa = 3.45FSGLTGTTISSINYY143 pKa = 9.57YY144 pKa = 10.76GGDD147 pKa = 3.63PSNGYY152 pKa = 10.06IIFANGEE159 pKa = 3.83RR160 pKa = 11.84MEE162 pKa = 4.03YY163 pKa = 10.28RR164 pKa = 11.84GIEE167 pKa = 4.67AITGTQDD174 pKa = 2.69GRR176 pKa = 11.84VDD178 pKa = 3.6GSTGADD184 pKa = 3.63SIGDD188 pKa = 4.21GYY190 pKa = 9.7TDD192 pKa = 3.56GQLDD196 pKa = 3.83AVGDD200 pKa = 4.16ADD202 pKa = 4.67GSHH205 pKa = 6.74NDD207 pKa = 3.18SIYY210 pKa = 10.73AYY212 pKa = 10.12GGNDD216 pKa = 3.2TVASGAGNDD225 pKa = 3.77TVYY228 pKa = 11.22GGAGTDD234 pKa = 3.35ALYY237 pKa = 11.06GGAGNDD243 pKa = 3.78ALDD246 pKa = 4.39GGSGDD251 pKa = 4.34DD252 pKa = 4.0ALSGGSGDD260 pKa = 3.7DD261 pKa = 3.31VLYY264 pKa = 11.0GGEE267 pKa = 4.3GWDD270 pKa = 3.49TLYY273 pKa = 11.23SGLGNDD279 pKa = 3.72TYY281 pKa = 11.71YY282 pKa = 11.29GGTGLDD288 pKa = 3.78YY289 pKa = 10.86IDD291 pKa = 5.7FGAEE295 pKa = 3.61TGGVYY300 pKa = 10.47VDD302 pKa = 5.0LSNGTIGGAATGDD315 pKa = 3.73VMGSGIDD322 pKa = 3.6GVFGSNHH329 pKa = 7.59DD330 pKa = 3.69DD331 pKa = 3.47TLIGFDD337 pKa = 3.54PWSPTGDD344 pKa = 3.01VYY346 pKa = 11.68TNVFFGRR353 pKa = 11.84AGNDD357 pKa = 3.1VFDD360 pKa = 4.53GRR362 pKa = 11.84GSPDD366 pKa = 3.39YY367 pKa = 10.23MDD369 pKa = 5.04GGSGDD374 pKa = 4.12DD375 pKa = 3.58TFLMTGTPGSDD386 pKa = 3.3TIIGGEE392 pKa = 4.2GGSDD396 pKa = 2.87QDD398 pKa = 4.23VIDD401 pKa = 4.57FSSVSGPITVDD412 pKa = 3.16YY413 pKa = 10.19TGAEE417 pKa = 4.22AGTITHH423 pKa = 6.83GADD426 pKa = 2.6TVTFSQIEE434 pKa = 4.27RR435 pKa = 11.84IYY437 pKa = 9.69ATSSGDD443 pKa = 3.62SLTGGAGTDD452 pKa = 3.1RR453 pKa = 11.84VEE455 pKa = 4.13GRR457 pKa = 11.84GGSDD461 pKa = 2.91TLYY464 pKa = 10.95GGAGGDD470 pKa = 3.89EE471 pKa = 4.39LSGDD475 pKa = 3.87SGDD478 pKa = 3.82DD479 pKa = 3.31SLYY482 pKa = 11.13GGTGADD488 pKa = 3.24TLYY491 pKa = 11.15GGLGADD497 pKa = 4.22RR498 pKa = 11.84IEE500 pKa = 4.84AGSGDD505 pKa = 3.61DD506 pKa = 4.35QIYY509 pKa = 10.68GGAGADD515 pKa = 4.1AIYY518 pKa = 10.36AGQGADD524 pKa = 3.51SIYY527 pKa = 10.91AASGDD532 pKa = 3.63DD533 pKa = 4.01AIYY536 pKa = 10.36GGSGSDD542 pKa = 3.39VVTLTGPGNYY552 pKa = 10.25ALFGGEE558 pKa = 4.18DD559 pKa = 3.44AGDD562 pKa = 3.86ADD564 pKa = 5.04VDD566 pKa = 3.98VLDD569 pKa = 4.71ISSLGAGGQVTAITYY584 pKa = 10.43NSGDD588 pKa = 4.25DD589 pKa = 3.57EE590 pKa = 5.42SGTITFSNGDD600 pKa = 3.27VATFSGFEE608 pKa = 4.14MIVCFAQGTRR618 pKa = 11.84IDD620 pKa = 3.9TPRR623 pKa = 11.84GPVAVEE629 pKa = 3.67DD630 pKa = 4.54LRR632 pKa = 11.84PGDD635 pKa = 3.9AVMTADD641 pKa = 5.34HH642 pKa = 6.92GPQPLRR648 pKa = 11.84WTGSRR653 pKa = 11.84RR654 pKa = 11.84VPARR658 pKa = 11.84GGSAPVTFATGAIGNTRR675 pKa = 11.84PLTLSPQHH683 pKa = 6.31RR684 pKa = 11.84VLITGWKK691 pKa = 8.98PEE693 pKa = 3.97MLFGEE698 pKa = 4.6AEE700 pKa = 4.14VLVAAKK706 pKa = 10.27HH707 pKa = 5.73LVDD710 pKa = 3.53GHH712 pKa = 6.27EE713 pKa = 4.5VTQSEE718 pKa = 5.03GSQITYY724 pKa = 9.89HH725 pKa = 6.28HH726 pKa = 7.11LCLDD730 pKa = 3.22RR731 pKa = 11.84HH732 pKa = 5.52EE733 pKa = 4.51VLFAEE738 pKa = 5.35GAPVEE743 pKa = 4.42SFLPAEE749 pKa = 4.22EE750 pKa = 4.74ALFSLTPAQRR760 pKa = 11.84RR761 pKa = 11.84DD762 pKa = 3.2LCAALPDD769 pKa = 3.9LDD771 pKa = 4.01AAARR775 pKa = 11.84PARR778 pKa = 11.84PILRR782 pKa = 11.84GYY784 pKa = 8.28EE785 pKa = 3.94AQLLGPSLPPPDD797 pKa = 4.26LRR799 pKa = 11.84RR800 pKa = 11.84AGHH803 pKa = 6.38RR804 pKa = 11.84MANRR808 pKa = 11.84ILL810 pKa = 3.66
MM1 pKa = 8.02ADD3 pKa = 3.23GKK5 pKa = 11.19YY6 pKa = 10.22DD7 pKa = 3.6GSSGSDD13 pKa = 3.09NINRR17 pKa = 11.84TSVDD21 pKa = 3.09GGGDD25 pKa = 3.42QVDD28 pKa = 4.62GPDD31 pKa = 3.74GLNDD35 pKa = 3.89SIYY38 pKa = 10.76GYY40 pKa = 10.56GGNDD44 pKa = 3.33TIEE47 pKa = 4.37AGRR50 pKa = 11.84GSDD53 pKa = 3.01TVYY56 pKa = 10.78GGEE59 pKa = 4.19GADD62 pKa = 3.76SIEE65 pKa = 4.16GRR67 pKa = 11.84SGDD70 pKa = 3.61DD71 pKa = 3.16VLYY74 pKa = 11.02GGAGRR79 pKa = 11.84DD80 pKa = 3.9TIEE83 pKa = 4.66GDD85 pKa = 3.19AGNDD89 pKa = 3.6TIYY92 pKa = 10.96VGSGDD97 pKa = 3.82DD98 pKa = 4.52DD99 pKa = 4.26IYY101 pKa = 11.64GEE103 pKa = 4.68SGDD106 pKa = 3.95DD107 pKa = 4.13LIRR110 pKa = 11.84VTRR113 pKa = 11.84GTSAWIDD120 pKa = 3.73GQSGDD125 pKa = 4.07DD126 pKa = 3.54TLDD129 pKa = 3.45FSGLTGTTISSINYY143 pKa = 9.57YY144 pKa = 10.76GGDD147 pKa = 3.63PSNGYY152 pKa = 10.06IIFANGEE159 pKa = 3.83RR160 pKa = 11.84MEE162 pKa = 4.03YY163 pKa = 10.28RR164 pKa = 11.84GIEE167 pKa = 4.67AITGTQDD174 pKa = 2.69GRR176 pKa = 11.84VDD178 pKa = 3.6GSTGADD184 pKa = 3.63SIGDD188 pKa = 4.21GYY190 pKa = 9.7TDD192 pKa = 3.56GQLDD196 pKa = 3.83AVGDD200 pKa = 4.16ADD202 pKa = 4.67GSHH205 pKa = 6.74NDD207 pKa = 3.18SIYY210 pKa = 10.73AYY212 pKa = 10.12GGNDD216 pKa = 3.2TVASGAGNDD225 pKa = 3.77TVYY228 pKa = 11.22GGAGTDD234 pKa = 3.35ALYY237 pKa = 11.06GGAGNDD243 pKa = 3.78ALDD246 pKa = 4.39GGSGDD251 pKa = 4.34DD252 pKa = 4.0ALSGGSGDD260 pKa = 3.7DD261 pKa = 3.31VLYY264 pKa = 11.0GGEE267 pKa = 4.3GWDD270 pKa = 3.49TLYY273 pKa = 11.23SGLGNDD279 pKa = 3.72TYY281 pKa = 11.71YY282 pKa = 11.29GGTGLDD288 pKa = 3.78YY289 pKa = 10.86IDD291 pKa = 5.7FGAEE295 pKa = 3.61TGGVYY300 pKa = 10.47VDD302 pKa = 5.0LSNGTIGGAATGDD315 pKa = 3.73VMGSGIDD322 pKa = 3.6GVFGSNHH329 pKa = 7.59DD330 pKa = 3.69DD331 pKa = 3.47TLIGFDD337 pKa = 3.54PWSPTGDD344 pKa = 3.01VYY346 pKa = 11.68TNVFFGRR353 pKa = 11.84AGNDD357 pKa = 3.1VFDD360 pKa = 4.53GRR362 pKa = 11.84GSPDD366 pKa = 3.39YY367 pKa = 10.23MDD369 pKa = 5.04GGSGDD374 pKa = 4.12DD375 pKa = 3.58TFLMTGTPGSDD386 pKa = 3.3TIIGGEE392 pKa = 4.2GGSDD396 pKa = 2.87QDD398 pKa = 4.23VIDD401 pKa = 4.57FSSVSGPITVDD412 pKa = 3.16YY413 pKa = 10.19TGAEE417 pKa = 4.22AGTITHH423 pKa = 6.83GADD426 pKa = 2.6TVTFSQIEE434 pKa = 4.27RR435 pKa = 11.84IYY437 pKa = 9.69ATSSGDD443 pKa = 3.62SLTGGAGTDD452 pKa = 3.1RR453 pKa = 11.84VEE455 pKa = 4.13GRR457 pKa = 11.84GGSDD461 pKa = 2.91TLYY464 pKa = 10.95GGAGGDD470 pKa = 3.89EE471 pKa = 4.39LSGDD475 pKa = 3.87SGDD478 pKa = 3.82DD479 pKa = 3.31SLYY482 pKa = 11.13GGTGADD488 pKa = 3.24TLYY491 pKa = 11.15GGLGADD497 pKa = 4.22RR498 pKa = 11.84IEE500 pKa = 4.84AGSGDD505 pKa = 3.61DD506 pKa = 4.35QIYY509 pKa = 10.68GGAGADD515 pKa = 4.1AIYY518 pKa = 10.36AGQGADD524 pKa = 3.51SIYY527 pKa = 10.91AASGDD532 pKa = 3.63DD533 pKa = 4.01AIYY536 pKa = 10.36GGSGSDD542 pKa = 3.39VVTLTGPGNYY552 pKa = 10.25ALFGGEE558 pKa = 4.18DD559 pKa = 3.44AGDD562 pKa = 3.86ADD564 pKa = 5.04VDD566 pKa = 3.98VLDD569 pKa = 4.71ISSLGAGGQVTAITYY584 pKa = 10.43NSGDD588 pKa = 4.25DD589 pKa = 3.57EE590 pKa = 5.42SGTITFSNGDD600 pKa = 3.27VATFSGFEE608 pKa = 4.14MIVCFAQGTRR618 pKa = 11.84IDD620 pKa = 3.9TPRR623 pKa = 11.84GPVAVEE629 pKa = 3.67DD630 pKa = 4.54LRR632 pKa = 11.84PGDD635 pKa = 3.9AVMTADD641 pKa = 5.34HH642 pKa = 6.92GPQPLRR648 pKa = 11.84WTGSRR653 pKa = 11.84RR654 pKa = 11.84VPARR658 pKa = 11.84GGSAPVTFATGAIGNTRR675 pKa = 11.84PLTLSPQHH683 pKa = 6.31RR684 pKa = 11.84VLITGWKK691 pKa = 8.98PEE693 pKa = 3.97MLFGEE698 pKa = 4.6AEE700 pKa = 4.14VLVAAKK706 pKa = 10.27HH707 pKa = 5.73LVDD710 pKa = 3.53GHH712 pKa = 6.27EE713 pKa = 4.5VTQSEE718 pKa = 5.03GSQITYY724 pKa = 9.89HH725 pKa = 6.28HH726 pKa = 7.11LCLDD730 pKa = 3.22RR731 pKa = 11.84HH732 pKa = 5.52EE733 pKa = 4.51VLFAEE738 pKa = 5.35GAPVEE743 pKa = 4.42SFLPAEE749 pKa = 4.22EE750 pKa = 4.74ALFSLTPAQRR760 pKa = 11.84RR761 pKa = 11.84DD762 pKa = 3.2LCAALPDD769 pKa = 3.9LDD771 pKa = 4.01AAARR775 pKa = 11.84PARR778 pKa = 11.84PILRR782 pKa = 11.84GYY784 pKa = 8.28EE785 pKa = 3.94AQLLGPSLPPPDD797 pKa = 4.26LRR799 pKa = 11.84RR800 pKa = 11.84AGHH803 pKa = 6.38RR804 pKa = 11.84MANRR808 pKa = 11.84ILL810 pKa = 3.66
Molecular weight: 82.12 kDa
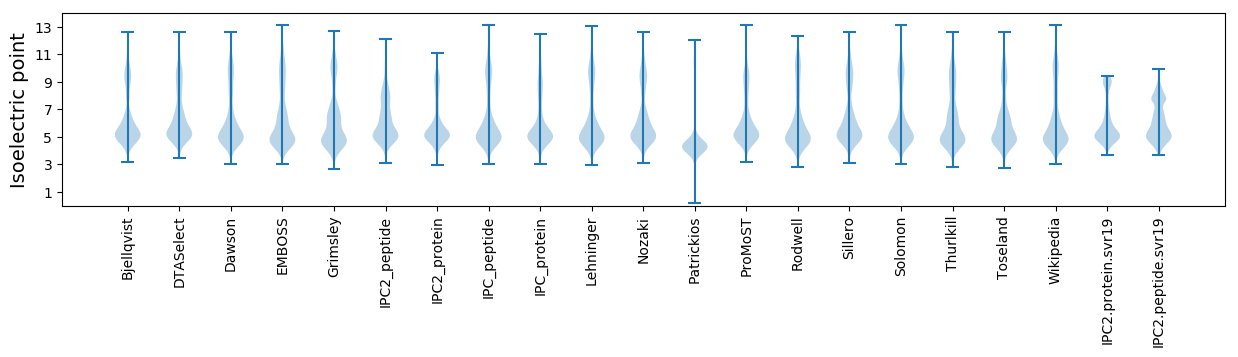
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I1JP25|A0A1I1JP25_9RHOB Chaperone required for the assembly of the F1-ATPase OS=Pseudooceanicola nitratireducens OX=517719 GN=SAMN05421762_1043 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1200732 |
30 |
2517 |
313.7 |
33.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.317 ± 0.055 | 0.852 ± 0.011 |
6.35 ± 0.039 | 5.701 ± 0.043 |
3.622 ± 0.023 | 8.824 ± 0.041 |
2.02 ± 0.02 | 5.137 ± 0.027 |
3.166 ± 0.032 | 10.063 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.814 ± 0.02 | 2.502 ± 0.023 |
5.184 ± 0.032 | 3.305 ± 0.025 |
6.712 ± 0.038 | 5.075 ± 0.028 |
5.577 ± 0.03 | 7.238 ± 0.03 |
1.333 ± 0.017 | 2.21 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
