
Streptocarpus flower break virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
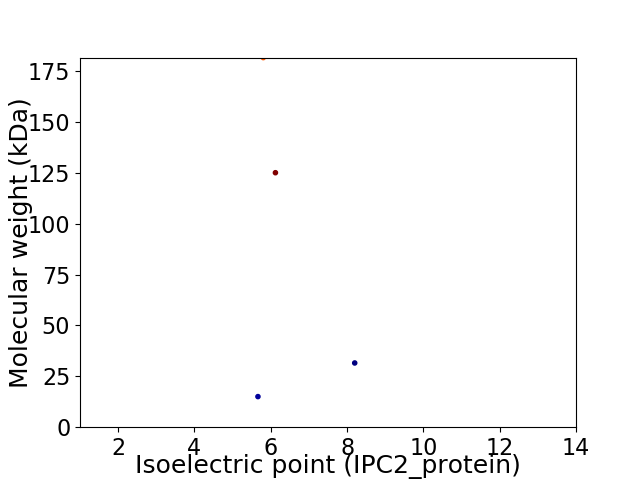
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1XF43|Q1XF43_9VIRU Movement protein OS=Streptocarpus flower break virus OX=335187 GN=p31 PE=3 SV=1
MM1 pKa = 7.47MNLCSASLSQIFQTQAARR19 pKa = 11.84DD20 pKa = 3.66AVRR23 pKa = 11.84EE24 pKa = 4.02QFGNTLRR31 pKa = 11.84SVVTQTTRR39 pKa = 11.84FPEE42 pKa = 4.29AGFQVYY48 pKa = 10.48INDD51 pKa = 4.07AALKK55 pKa = 9.17PLWEE59 pKa = 4.38NLLKK63 pKa = 11.11SFDD66 pKa = 3.61TKK68 pKa = 11.27NRR70 pKa = 11.84ILEE73 pKa = 4.43TEE75 pKa = 4.12EE76 pKa = 3.49EE77 pKa = 4.54TRR79 pKa = 11.84PSTAEE84 pKa = 3.77VLNATQRR91 pKa = 11.84VDD93 pKa = 4.05DD94 pKa = 3.98STTAIRR100 pKa = 11.84GALQRR105 pKa = 11.84LSDD108 pKa = 3.73EE109 pKa = 4.6LNRR112 pKa = 11.84GSGYY116 pKa = 8.26MNRR119 pKa = 11.84TSFEE123 pKa = 4.41TILLWSAATAKK134 pKa = 10.65
MM1 pKa = 7.47MNLCSASLSQIFQTQAARR19 pKa = 11.84DD20 pKa = 3.66AVRR23 pKa = 11.84EE24 pKa = 4.02QFGNTLRR31 pKa = 11.84SVVTQTTRR39 pKa = 11.84FPEE42 pKa = 4.29AGFQVYY48 pKa = 10.48INDD51 pKa = 4.07AALKK55 pKa = 9.17PLWEE59 pKa = 4.38NLLKK63 pKa = 11.11SFDD66 pKa = 3.61TKK68 pKa = 11.27NRR70 pKa = 11.84ILEE73 pKa = 4.43TEE75 pKa = 4.12EE76 pKa = 3.49EE77 pKa = 4.54TRR79 pKa = 11.84PSTAEE84 pKa = 3.77VLNATQRR91 pKa = 11.84VDD93 pKa = 4.05DD94 pKa = 3.98STTAIRR100 pKa = 11.84GALQRR105 pKa = 11.84LSDD108 pKa = 3.73EE109 pKa = 4.6LNRR112 pKa = 11.84GSGYY116 pKa = 8.26MNRR119 pKa = 11.84TSFEE123 pKa = 4.41TILLWSAATAKK134 pKa = 10.65
Molecular weight: 15.04 kDa
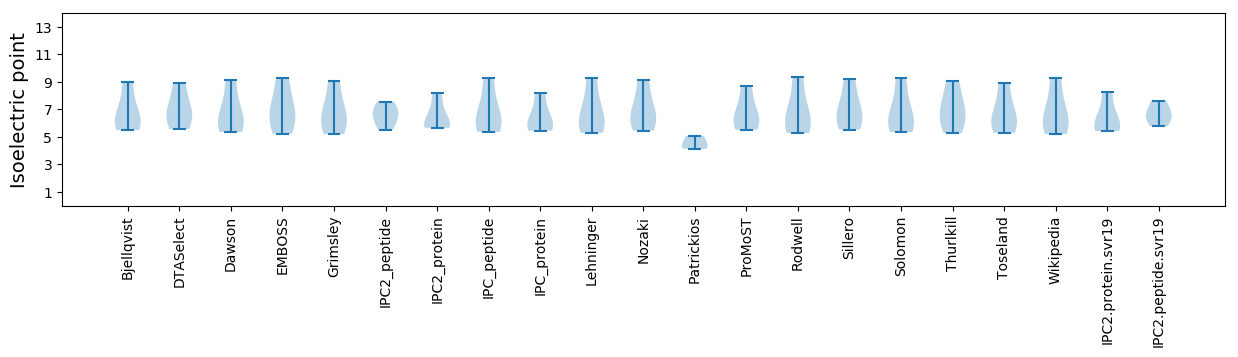
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1XF44|Q1XF44_9VIRU Methyltransferase/RNA helicase OS=Streptocarpus flower break virus OX=335187 GN=55 PE=4 SV=1
MM1 pKa = 7.7AIVLSKK7 pKa = 10.73PKK9 pKa = 10.41VSEE12 pKa = 3.86FLNLTKK18 pKa = 10.55AEE20 pKa = 4.35EE21 pKa = 4.08ILPKK25 pKa = 10.38FLTRR29 pKa = 11.84LKK31 pKa = 9.59TVAISTRR38 pKa = 11.84DD39 pKa = 3.53VVSVKK44 pKa = 9.7GTTDD48 pKa = 3.37LVDD51 pKa = 3.46IDD53 pKa = 4.7LLRR56 pKa = 11.84DD57 pKa = 3.49VPVNNWRR64 pKa = 11.84YY65 pKa = 8.87VGIVGIVASGEE76 pKa = 3.76WLLPDD81 pKa = 3.65NVSGGVAISFVDD93 pKa = 3.22KK94 pKa = 10.83RR95 pKa = 11.84LVDD98 pKa = 3.55SRR100 pKa = 11.84EE101 pKa = 4.45AILGTYY107 pKa = 9.34RR108 pKa = 11.84AAAVEE113 pKa = 4.04KK114 pKa = 10.27RR115 pKa = 11.84FQFKK119 pKa = 10.75LIPNYY124 pKa = 9.94FVSQEE129 pKa = 3.94DD130 pKa = 3.49ALRR133 pKa = 11.84RR134 pKa = 11.84PWQVQVSLKK143 pKa = 10.35GLKK146 pKa = 10.02FEE148 pKa = 5.12EE149 pKa = 5.17GFSPLTLEE157 pKa = 4.12FVSVVVCANSVVTKK171 pKa = 9.92GLRR174 pKa = 11.84EE175 pKa = 3.83RR176 pKa = 11.84LNNVGDD182 pKa = 4.04PNVEE186 pKa = 3.82VSEE189 pKa = 4.48VVVDD193 pKa = 3.89EE194 pKa = 4.7FVDD197 pKa = 4.48SISASQSLSRR207 pKa = 11.84ARR209 pKa = 11.84NKK211 pKa = 9.96YY212 pKa = 9.06VRR214 pKa = 11.84GNGKK218 pKa = 9.57VGNNSGGFSKK228 pKa = 10.94YY229 pKa = 9.77NRR231 pKa = 11.84HH232 pKa = 4.71QPEE235 pKa = 4.08RR236 pKa = 11.84FAGKK240 pKa = 10.53AMYY243 pKa = 9.98NSKK246 pKa = 10.61NVVRR250 pKa = 11.84GGTSEE255 pKa = 3.88PTAILHH261 pKa = 6.1KK262 pKa = 10.8RR263 pKa = 11.84MGGSNSIDD271 pKa = 3.06EE272 pKa = 4.67SLFSVAEE279 pKa = 4.05SDD281 pKa = 5.33LSDD284 pKa = 3.73SGCAA288 pKa = 3.53
MM1 pKa = 7.7AIVLSKK7 pKa = 10.73PKK9 pKa = 10.41VSEE12 pKa = 3.86FLNLTKK18 pKa = 10.55AEE20 pKa = 4.35EE21 pKa = 4.08ILPKK25 pKa = 10.38FLTRR29 pKa = 11.84LKK31 pKa = 9.59TVAISTRR38 pKa = 11.84DD39 pKa = 3.53VVSVKK44 pKa = 9.7GTTDD48 pKa = 3.37LVDD51 pKa = 3.46IDD53 pKa = 4.7LLRR56 pKa = 11.84DD57 pKa = 3.49VPVNNWRR64 pKa = 11.84YY65 pKa = 8.87VGIVGIVASGEE76 pKa = 3.76WLLPDD81 pKa = 3.65NVSGGVAISFVDD93 pKa = 3.22KK94 pKa = 10.83RR95 pKa = 11.84LVDD98 pKa = 3.55SRR100 pKa = 11.84EE101 pKa = 4.45AILGTYY107 pKa = 9.34RR108 pKa = 11.84AAAVEE113 pKa = 4.04KK114 pKa = 10.27RR115 pKa = 11.84FQFKK119 pKa = 10.75LIPNYY124 pKa = 9.94FVSQEE129 pKa = 3.94DD130 pKa = 3.49ALRR133 pKa = 11.84RR134 pKa = 11.84PWQVQVSLKK143 pKa = 10.35GLKK146 pKa = 10.02FEE148 pKa = 5.12EE149 pKa = 5.17GFSPLTLEE157 pKa = 4.12FVSVVVCANSVVTKK171 pKa = 9.92GLRR174 pKa = 11.84EE175 pKa = 3.83RR176 pKa = 11.84LNNVGDD182 pKa = 4.04PNVEE186 pKa = 3.82VSEE189 pKa = 4.48VVVDD193 pKa = 3.89EE194 pKa = 4.7FVDD197 pKa = 4.48SISASQSLSRR207 pKa = 11.84ARR209 pKa = 11.84NKK211 pKa = 9.96YY212 pKa = 9.06VRR214 pKa = 11.84GNGKK218 pKa = 9.57VGNNSGGFSKK228 pKa = 10.94YY229 pKa = 9.77NRR231 pKa = 11.84HH232 pKa = 4.71QPEE235 pKa = 4.08RR236 pKa = 11.84FAGKK240 pKa = 10.53AMYY243 pKa = 9.98NSKK246 pKa = 10.61NVVRR250 pKa = 11.84GGTSEE255 pKa = 3.88PTAILHH261 pKa = 6.1KK262 pKa = 10.8RR263 pKa = 11.84MGGSNSIDD271 pKa = 3.06EE272 pKa = 4.67SLFSVAEE279 pKa = 4.05SDD281 pKa = 5.33LSDD284 pKa = 3.73SGCAA288 pKa = 3.53
Molecular weight: 31.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3133 |
134 |
1606 |
783.3 |
88.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.767 ± 0.467 | 1.564 ± 0.193 |
6.096 ± 0.306 | 6.192 ± 0.21 |
5.235 ± 0.201 | 4.915 ± 0.407 |
2.266 ± 0.414 | 4.692 ± 0.39 |
6.575 ± 0.48 | 9.863 ± 0.187 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.947 ± 0.136 | 4.852 ± 0.251 |
3.671 ± 0.199 | 3.288 ± 0.356 |
5.011 ± 0.532 | 7.533 ± 0.458 |
5.905 ± 0.703 | 9.129 ± 0.887 |
0.989 ± 0.071 | 3.479 ± 0.377 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
