
Microbacterium sp. BK668
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
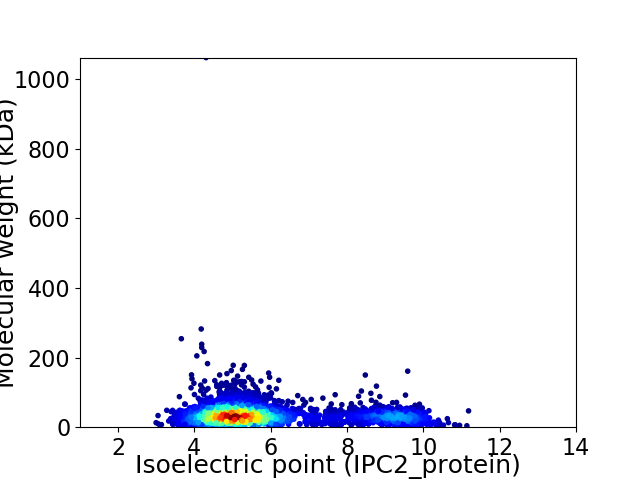
Virtual 2D-PAGE plot for 3319 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
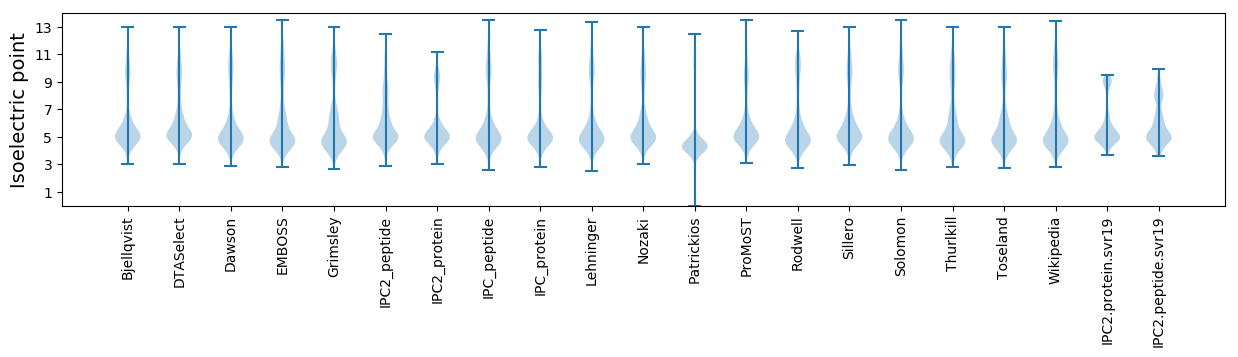
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4R6G9S2|A0A4R6G9S2_9MICO Carbohydrate ABC transporter membrane protein 2 (CUT1 family) OS=Microbacterium sp. BK668 OX=2512118 GN=EV279_0950 PE=3 SV=1
MM1 pKa = 7.37RR2 pKa = 11.84HH3 pKa = 5.09RR4 pKa = 11.84WYY6 pKa = 10.54AGAVATLVSGALIFSGVGPAVAAEE30 pKa = 4.25VTPTPTPEE38 pKa = 3.43QSAAPEE44 pKa = 3.78QTAAPEE50 pKa = 4.6DD51 pKa = 4.24PTDD54 pKa = 4.24PPADD58 pKa = 3.54TPIQEE63 pKa = 4.28EE64 pKa = 4.73SPAPEE69 pKa = 5.03DD70 pKa = 3.7PAPADD75 pKa = 3.96PEE77 pKa = 4.26PSEE80 pKa = 4.11PAAPGGDD87 pKa = 3.78VKK89 pKa = 10.93PADD92 pKa = 3.65QTDD95 pKa = 3.85LSVQDD100 pKa = 3.97EE101 pKa = 4.5QKK103 pKa = 9.57TAQDD107 pKa = 3.21AAAAQPDD114 pKa = 4.21VGVLLVPDD122 pKa = 5.42PGPTTSVITVKK133 pKa = 10.65VGSDD137 pKa = 2.87RR138 pKa = 11.84TGITGVTSLAGVVLLLNTGGAGGPSGTRR166 pKa = 11.84PDD168 pKa = 3.91GVAGTGDD175 pKa = 3.33GWAKK179 pKa = 9.12CTSDD183 pKa = 3.64AQGDD187 pKa = 4.05CSFTVPATNPGGANRR202 pKa = 11.84DD203 pKa = 3.01QRR205 pKa = 11.84YY206 pKa = 7.56WVVQSSVPAGYY217 pKa = 9.7YY218 pKa = 10.64ANPTLRR224 pKa = 11.84TGGSSGAGAATPYY237 pKa = 9.97TFRR240 pKa = 11.84TGNQLRR246 pKa = 11.84ANSTYY251 pKa = 10.96SSQDD255 pKa = 2.58ANAFMLSSGITPTASGGIWQQSRR278 pKa = 11.84SNPALTASCGLDD290 pKa = 3.05VALILDD296 pKa = 4.2LSGSVSGSLAQLKK309 pKa = 7.85TAANTFVDD317 pKa = 4.22SLQGTPSRR325 pKa = 11.84MALFSFSSQTPATGATANFPSLTPVSTPAQGTAFKK360 pKa = 10.81NLYY363 pKa = 10.11APWTADD369 pKa = 4.02GSTNWDD375 pKa = 3.3RR376 pKa = 11.84GLGVAAAANTAANNFDD392 pKa = 3.46IAVVITDD399 pKa = 4.59GNPTSYY405 pKa = 10.66NQPPQGSGSDD415 pKa = 3.16NRR417 pKa = 11.84FRR419 pKa = 11.84EE420 pKa = 4.25TEE422 pKa = 3.82NGIFSANALKK432 pKa = 10.64AGPGGTAPTRR442 pKa = 11.84VLAFGVGDD450 pKa = 3.97GATGATNALNLRR462 pKa = 11.84AISGPTAFTGANGEE476 pKa = 4.22TADD479 pKa = 4.15YY480 pKa = 10.32YY481 pKa = 10.38QANNFAAVGTALRR494 pKa = 11.84NLALGNCEE502 pKa = 3.77GTLTVTKK509 pKa = 10.17QIIPNTTPVGQTTGALPSGAGWQFTGTVNTPGVTTPAAARR549 pKa = 11.84TTTADD554 pKa = 3.04GTGTVTYY561 pKa = 9.86PLAFPGGTTSASVSVVEE578 pKa = 4.44TQQSGFTLQQVGGQNAVCRR597 pKa = 11.84NLNTNEE603 pKa = 3.91AVSVTNSGALGFTVDD618 pKa = 3.76VPSTQAVNCVVFNRR632 pKa = 11.84APAPEE637 pKa = 4.51ANLTVTKK644 pKa = 8.8TWVVNGVTYY653 pKa = 10.39PDD655 pKa = 4.02GAQPSDD661 pKa = 3.53LTAQLQLTGPGAAGATNQGWGVTRR685 pKa = 11.84TGYY688 pKa = 10.75AQGDD692 pKa = 3.88TTTLSEE698 pKa = 4.36QVTLIDD704 pKa = 3.64PTMCTSSAVVTSVNGTPANVPLGAGYY730 pKa = 10.93QMLLSQINNTATITNTVLCNSEE752 pKa = 3.93LTLIKK757 pKa = 10.14QVQGGDD763 pKa = 3.65AQPSSWTLNASFLATAQVPTGLPGFSGAAEE793 pKa = 4.23SASVASQNVTPNARR807 pKa = 11.84YY808 pKa = 9.34QLFEE812 pKa = 4.25SGGDD816 pKa = 3.26PRR818 pKa = 11.84YY819 pKa = 10.08RR820 pKa = 11.84QTDD823 pKa = 2.9NRR825 pKa = 11.84TNLQSNPLSTGSATCIRR842 pKa = 11.84VEE844 pKa = 4.34ADD846 pKa = 2.89GDD848 pKa = 3.69PWPGSGFSDD857 pKa = 5.34GINGGVNVPLGYY869 pKa = 9.94RR870 pKa = 11.84VACTLVNQTADD881 pKa = 3.4LTLLKK886 pKa = 9.97TVVNDD891 pKa = 3.81NGGSTPPSAWDD902 pKa = 3.48LTATPAVLAGLTPTTVPGSEE922 pKa = 4.11TAGVANTFGVRR933 pKa = 11.84PGHH936 pKa = 6.4VYY938 pKa = 9.89TLTEE942 pKa = 4.01SDD944 pKa = 3.67VPGYY948 pKa = 10.57QFAKK952 pKa = 10.04LQRR955 pKa = 11.84FVNGAWVDD963 pKa = 4.05VVADD967 pKa = 4.67PDD969 pKa = 3.99PLRR972 pKa = 11.84YY973 pKa = 8.6PQQNGDD979 pKa = 4.11GDD981 pKa = 3.92WLITVDD987 pKa = 4.63PLDD990 pKa = 4.35DD991 pKa = 4.05PLYY994 pKa = 10.87RR995 pKa = 11.84FVNDD999 pKa = 5.37DD1000 pKa = 3.44IAPQLTLVKK1009 pKa = 10.32EE1010 pKa = 4.27VTNDD1014 pKa = 3.14NGGEE1018 pKa = 4.02AEE1020 pKa = 4.19PTDD1023 pKa = 3.48WTLTATTPGGPDD1035 pKa = 3.2LSGQTEE1041 pKa = 4.6TPAVTAQPVRR1051 pKa = 11.84AGVVYY1056 pKa = 9.54TIGEE1060 pKa = 4.19NGGPAAYY1067 pKa = 10.13SLNSLTCTGYY1077 pKa = 10.11PNTSVGNPTITLAPGDD1093 pKa = 4.78DD1094 pKa = 4.04VTCTLVNDD1102 pKa = 5.56DD1103 pKa = 3.75ILIPVTVEE1111 pKa = 3.68KK1112 pKa = 11.02SDD1114 pKa = 3.61GTVEE1118 pKa = 3.92QLADD1122 pKa = 3.63GTWRR1126 pKa = 11.84ITYY1129 pKa = 9.13EE1130 pKa = 4.03VVVANGSPTLATRR1143 pKa = 11.84FSLTDD1148 pKa = 3.22TPQFDD1153 pKa = 3.6TSFTVLTQGWEE1164 pKa = 4.06GSPNVTDD1171 pKa = 3.83VPIEE1175 pKa = 4.2GGGVYY1180 pKa = 9.79TFTYY1184 pKa = 10.64VVTAEE1189 pKa = 4.11ANIEE1193 pKa = 4.02PVAPGALVCSPTNGGGFFNSATVTFPSGTDD1223 pKa = 3.04TDD1225 pKa = 3.95TGCAVPGAPTVQKK1238 pKa = 9.12TAQTAVQNPATGEE1251 pKa = 3.61WTLRR1255 pKa = 11.84YY1256 pKa = 9.19LVAVSNPTPIPLSYY1270 pKa = 10.07TLSDD1274 pKa = 3.35TAAPLPAGVAGGAWSASDD1292 pKa = 3.66PTPVGGGTFVRR1303 pKa = 11.84NAGWSGAGEE1312 pKa = 4.42LATGTLPAGAVHH1324 pKa = 7.0TYY1326 pKa = 10.58AVTRR1330 pKa = 11.84VVTVAASVSEE1340 pKa = 4.0AALTCGSTPADD1351 pKa = 3.95GGGVWNTASVTNGITVTEE1369 pKa = 4.38SSDD1372 pKa = 3.52CVDD1375 pKa = 3.24IEE1377 pKa = 4.36RR1378 pKa = 11.84PAVTIAKK1385 pKa = 8.14TVTDD1389 pKa = 3.87TRR1391 pKa = 11.84QLADD1395 pKa = 3.74GTWEE1399 pKa = 3.62IVYY1402 pKa = 10.44DD1403 pKa = 3.9VVVTNSSTTLAAVYY1417 pKa = 10.5SLADD1421 pKa = 3.46SLQFGGDD1428 pKa = 3.02ITVEE1432 pKa = 4.28GASWTGPTSGDD1443 pKa = 3.48FAPDD1447 pKa = 3.24GTAEE1451 pKa = 4.16LATDD1455 pKa = 3.9RR1456 pKa = 11.84VLAPLVGDD1464 pKa = 4.38AGVEE1468 pKa = 4.46TYY1470 pKa = 11.01VVTARR1475 pKa = 11.84ATVDD1479 pKa = 3.7LAAWEE1484 pKa = 4.79GDD1486 pKa = 3.66TLSCTDD1492 pKa = 4.54EE1493 pKa = 4.78DD1494 pKa = 4.73PPTAGGFLNVATVTVNGVGIPADD1517 pKa = 3.94DD1518 pKa = 4.18CSEE1521 pKa = 4.49PGLPTIAKK1529 pKa = 9.31QGVAATQDD1537 pKa = 3.54PSDD1540 pKa = 3.84PSAWTVTYY1548 pKa = 10.08EE1549 pKa = 4.04VTVTSGGFDD1558 pKa = 3.19TFYY1561 pKa = 11.31SLSDD1565 pKa = 3.51TPAFAAGIALNAGTAQRR1582 pKa = 11.84TDD1584 pKa = 3.1IADD1587 pKa = 3.67QPVLPISPGADD1598 pKa = 2.9FVTDD1602 pKa = 3.3VALAADD1608 pKa = 3.87ATHH1611 pKa = 6.7VYY1613 pKa = 9.74LVSWSVEE1620 pKa = 3.79VTDD1623 pKa = 6.14AFTEE1627 pKa = 4.27DD1628 pKa = 4.75DD1629 pKa = 4.21ADD1631 pKa = 4.0CTGEE1635 pKa = 4.08PGSGFFNSATLLVGDD1650 pKa = 4.68IPIDD1654 pKa = 3.94GSDD1657 pKa = 4.84CIPVADD1663 pKa = 4.1RR1664 pKa = 11.84VYY1666 pKa = 8.38PTIVKK1671 pKa = 8.67TVTGTDD1677 pKa = 3.43QDD1679 pKa = 4.23PSTGDD1684 pKa = 2.93WTITYY1689 pKa = 9.97AIEE1692 pKa = 3.9VTLAPVGPGNPDD1704 pKa = 2.99GLAAEE1709 pKa = 4.47YY1710 pKa = 11.04DD1711 pKa = 3.76LTDD1714 pKa = 3.48TLDD1717 pKa = 3.48FGGGIEE1723 pKa = 4.32VVEE1726 pKa = 4.34ATWAGQSSGTFDD1738 pKa = 4.04PVTDD1742 pKa = 4.05PAEE1745 pKa = 4.23LASDD1749 pKa = 3.59EE1750 pKa = 5.09AIAAGATHH1758 pKa = 6.94VYY1760 pKa = 10.18SVTVRR1765 pKa = 11.84ATVTAEE1771 pKa = 4.48AIDD1774 pKa = 4.57GGTTSCDD1781 pKa = 3.26PGEE1784 pKa = 4.37GQTGGFLNTALLSSGGEE1801 pKa = 4.02DD1802 pKa = 3.63TPVDD1806 pKa = 3.45ACAEE1810 pKa = 4.21PVFPTISKK1818 pKa = 10.58SSAQGGAVQQPDD1830 pKa = 4.26GSWALAYY1837 pKa = 9.9DD1838 pKa = 3.45ITVSYY1843 pKa = 10.03PSTDD1847 pKa = 3.34ADD1849 pKa = 3.55PRR1851 pKa = 11.84PSVGYY1856 pKa = 10.61VLTDD1860 pKa = 3.86VPDD1863 pKa = 4.06LPANVEE1869 pKa = 4.22LQGEE1873 pKa = 4.37WTATAGADD1881 pKa = 3.73TPAPDD1886 pKa = 5.16DD1887 pKa = 3.59PTFDD1891 pKa = 5.3GEE1893 pKa = 4.71GTWTIVTAALDD1904 pKa = 3.92PDD1906 pKa = 3.94EE1907 pKa = 6.62DD1908 pKa = 4.25GVTQHH1913 pKa = 6.23VYY1915 pKa = 10.28TITAVVRR1922 pKa = 11.84VTAPPVEE1929 pKa = 4.38PVAEE1933 pKa = 4.09CAEE1936 pKa = 4.19LEE1938 pKa = 4.18EE1939 pKa = 4.42TGIVVPNSATVTSGGYY1955 pKa = 7.58TADD1958 pKa = 4.84DD1959 pKa = 4.46DD1960 pKa = 4.27ACQVVQFDD1968 pKa = 4.08DD1969 pKa = 3.83VGVEE1973 pKa = 4.12KK1974 pKa = 10.88SADD1977 pKa = 3.48LAGEE1981 pKa = 4.21EE1982 pKa = 4.5TSVEE1986 pKa = 4.03PGDD1989 pKa = 3.51QFDD1992 pKa = 4.1YY1993 pKa = 11.35VITVTNTGTGPANEE2007 pKa = 4.28ILVFDD2012 pKa = 5.3DD2013 pKa = 4.42SLSEE2017 pKa = 3.84APYY2020 pKa = 9.6VGRR2023 pKa = 11.84VQLVAGTLSVEE2034 pKa = 4.3PAGLAYY2040 pKa = 9.49TDD2042 pKa = 4.66SSDD2045 pKa = 4.43LPGNVIDD2052 pKa = 6.39LEE2054 pKa = 4.16IDD2056 pKa = 3.25QLAVGEE2062 pKa = 4.43SATIRR2067 pKa = 11.84VTAEE2071 pKa = 3.61FLPAPVGGEE2080 pKa = 3.82PQVPPGEE2087 pKa = 4.92DD2088 pKa = 3.52PPAPTPPLDD2097 pKa = 4.75AIVNLVCVAAEE2108 pKa = 3.74FDD2110 pKa = 4.29GNPEE2114 pKa = 4.08NNCDD2118 pKa = 3.87DD2119 pKa = 4.71AEE2121 pKa = 4.38VPTRR2125 pKa = 11.84DD2126 pKa = 2.94ITASVYY2132 pKa = 8.46ATCQSDD2138 pKa = 3.87APLLGWVVAKK2148 pKa = 10.49SALLAGEE2155 pKa = 4.2PASLLWEE2162 pKa = 5.09LDD2164 pKa = 3.5PPSPGSVPPFVEE2176 pKa = 4.37FDD2178 pKa = 3.47EE2179 pKa = 4.9SDD2181 pKa = 3.35EE2182 pKa = 4.44GAVGPVWTGLADD2194 pKa = 3.32WPGAFFTPSGVAIDD2208 pKa = 4.02YY2209 pKa = 8.49PGWRR2213 pKa = 11.84PIEE2216 pKa = 4.26LSDD2219 pKa = 4.44VVPGSNPINYY2229 pKa = 7.99YY2230 pKa = 10.08IPGPEE2235 pKa = 4.41PRR2237 pKa = 11.84TEE2239 pKa = 3.58MTAADD2244 pKa = 4.62RR2245 pKa = 11.84LNLVYY2250 pKa = 10.76NGLILDD2256 pKa = 4.48DD2257 pKa = 5.05SEE2259 pKa = 6.32LDD2261 pKa = 3.86YY2262 pKa = 11.4AWRR2265 pKa = 11.84GASTVTLSVNPEE2277 pKa = 3.62LTFEE2281 pKa = 4.23VEE2283 pKa = 4.42YY2284 pKa = 10.46PPATPEE2290 pKa = 4.12CVQARR2295 pKa = 11.84HH2296 pKa = 5.0STVEE2300 pKa = 3.77IEE2302 pKa = 4.08KK2303 pKa = 8.62TASVEE2308 pKa = 4.02RR2309 pKa = 11.84TSAGQSFTYY2318 pKa = 10.17SLEE2321 pKa = 4.15VANVSDD2327 pKa = 4.49DD2328 pKa = 3.76SAAEE2332 pKa = 3.87AVVITDD2338 pKa = 5.83AIPADD2343 pKa = 3.83LRR2345 pKa = 11.84ITSVTWPGQGDD2356 pKa = 3.56AGVFPNWSSCAVTGQNAQGYY2376 pKa = 9.53GGVLNCTLFGPLQPVGANDD2395 pKa = 4.05SPSSAPTITLAATVNPASTAAVITNVAVVEE2425 pKa = 4.21YY2426 pKa = 8.68QTFGDD2431 pKa = 4.31PEE2433 pKa = 4.24DD2434 pKa = 4.09TGTDD2438 pKa = 3.39SDD2440 pKa = 4.5DD2441 pKa = 3.58ATVLLSGLPVTGGSPALPLIMLGFLALLGGVATLVVTRR2479 pKa = 11.84RR2480 pKa = 11.84RR2481 pKa = 11.84RR2482 pKa = 11.84KK2483 pKa = 9.1SATVDD2488 pKa = 3.4VQQ2490 pKa = 3.25
MM1 pKa = 7.37RR2 pKa = 11.84HH3 pKa = 5.09RR4 pKa = 11.84WYY6 pKa = 10.54AGAVATLVSGALIFSGVGPAVAAEE30 pKa = 4.25VTPTPTPEE38 pKa = 3.43QSAAPEE44 pKa = 3.78QTAAPEE50 pKa = 4.6DD51 pKa = 4.24PTDD54 pKa = 4.24PPADD58 pKa = 3.54TPIQEE63 pKa = 4.28EE64 pKa = 4.73SPAPEE69 pKa = 5.03DD70 pKa = 3.7PAPADD75 pKa = 3.96PEE77 pKa = 4.26PSEE80 pKa = 4.11PAAPGGDD87 pKa = 3.78VKK89 pKa = 10.93PADD92 pKa = 3.65QTDD95 pKa = 3.85LSVQDD100 pKa = 3.97EE101 pKa = 4.5QKK103 pKa = 9.57TAQDD107 pKa = 3.21AAAAQPDD114 pKa = 4.21VGVLLVPDD122 pKa = 5.42PGPTTSVITVKK133 pKa = 10.65VGSDD137 pKa = 2.87RR138 pKa = 11.84TGITGVTSLAGVVLLLNTGGAGGPSGTRR166 pKa = 11.84PDD168 pKa = 3.91GVAGTGDD175 pKa = 3.33GWAKK179 pKa = 9.12CTSDD183 pKa = 3.64AQGDD187 pKa = 4.05CSFTVPATNPGGANRR202 pKa = 11.84DD203 pKa = 3.01QRR205 pKa = 11.84YY206 pKa = 7.56WVVQSSVPAGYY217 pKa = 9.7YY218 pKa = 10.64ANPTLRR224 pKa = 11.84TGGSSGAGAATPYY237 pKa = 9.97TFRR240 pKa = 11.84TGNQLRR246 pKa = 11.84ANSTYY251 pKa = 10.96SSQDD255 pKa = 2.58ANAFMLSSGITPTASGGIWQQSRR278 pKa = 11.84SNPALTASCGLDD290 pKa = 3.05VALILDD296 pKa = 4.2LSGSVSGSLAQLKK309 pKa = 7.85TAANTFVDD317 pKa = 4.22SLQGTPSRR325 pKa = 11.84MALFSFSSQTPATGATANFPSLTPVSTPAQGTAFKK360 pKa = 10.81NLYY363 pKa = 10.11APWTADD369 pKa = 4.02GSTNWDD375 pKa = 3.3RR376 pKa = 11.84GLGVAAAANTAANNFDD392 pKa = 3.46IAVVITDD399 pKa = 4.59GNPTSYY405 pKa = 10.66NQPPQGSGSDD415 pKa = 3.16NRR417 pKa = 11.84FRR419 pKa = 11.84EE420 pKa = 4.25TEE422 pKa = 3.82NGIFSANALKK432 pKa = 10.64AGPGGTAPTRR442 pKa = 11.84VLAFGVGDD450 pKa = 3.97GATGATNALNLRR462 pKa = 11.84AISGPTAFTGANGEE476 pKa = 4.22TADD479 pKa = 4.15YY480 pKa = 10.32YY481 pKa = 10.38QANNFAAVGTALRR494 pKa = 11.84NLALGNCEE502 pKa = 3.77GTLTVTKK509 pKa = 10.17QIIPNTTPVGQTTGALPSGAGWQFTGTVNTPGVTTPAAARR549 pKa = 11.84TTTADD554 pKa = 3.04GTGTVTYY561 pKa = 9.86PLAFPGGTTSASVSVVEE578 pKa = 4.44TQQSGFTLQQVGGQNAVCRR597 pKa = 11.84NLNTNEE603 pKa = 3.91AVSVTNSGALGFTVDD618 pKa = 3.76VPSTQAVNCVVFNRR632 pKa = 11.84APAPEE637 pKa = 4.51ANLTVTKK644 pKa = 8.8TWVVNGVTYY653 pKa = 10.39PDD655 pKa = 4.02GAQPSDD661 pKa = 3.53LTAQLQLTGPGAAGATNQGWGVTRR685 pKa = 11.84TGYY688 pKa = 10.75AQGDD692 pKa = 3.88TTTLSEE698 pKa = 4.36QVTLIDD704 pKa = 3.64PTMCTSSAVVTSVNGTPANVPLGAGYY730 pKa = 10.93QMLLSQINNTATITNTVLCNSEE752 pKa = 3.93LTLIKK757 pKa = 10.14QVQGGDD763 pKa = 3.65AQPSSWTLNASFLATAQVPTGLPGFSGAAEE793 pKa = 4.23SASVASQNVTPNARR807 pKa = 11.84YY808 pKa = 9.34QLFEE812 pKa = 4.25SGGDD816 pKa = 3.26PRR818 pKa = 11.84YY819 pKa = 10.08RR820 pKa = 11.84QTDD823 pKa = 2.9NRR825 pKa = 11.84TNLQSNPLSTGSATCIRR842 pKa = 11.84VEE844 pKa = 4.34ADD846 pKa = 2.89GDD848 pKa = 3.69PWPGSGFSDD857 pKa = 5.34GINGGVNVPLGYY869 pKa = 9.94RR870 pKa = 11.84VACTLVNQTADD881 pKa = 3.4LTLLKK886 pKa = 9.97TVVNDD891 pKa = 3.81NGGSTPPSAWDD902 pKa = 3.48LTATPAVLAGLTPTTVPGSEE922 pKa = 4.11TAGVANTFGVRR933 pKa = 11.84PGHH936 pKa = 6.4VYY938 pKa = 9.89TLTEE942 pKa = 4.01SDD944 pKa = 3.67VPGYY948 pKa = 10.57QFAKK952 pKa = 10.04LQRR955 pKa = 11.84FVNGAWVDD963 pKa = 4.05VVADD967 pKa = 4.67PDD969 pKa = 3.99PLRR972 pKa = 11.84YY973 pKa = 8.6PQQNGDD979 pKa = 4.11GDD981 pKa = 3.92WLITVDD987 pKa = 4.63PLDD990 pKa = 4.35DD991 pKa = 4.05PLYY994 pKa = 10.87RR995 pKa = 11.84FVNDD999 pKa = 5.37DD1000 pKa = 3.44IAPQLTLVKK1009 pKa = 10.32EE1010 pKa = 4.27VTNDD1014 pKa = 3.14NGGEE1018 pKa = 4.02AEE1020 pKa = 4.19PTDD1023 pKa = 3.48WTLTATTPGGPDD1035 pKa = 3.2LSGQTEE1041 pKa = 4.6TPAVTAQPVRR1051 pKa = 11.84AGVVYY1056 pKa = 9.54TIGEE1060 pKa = 4.19NGGPAAYY1067 pKa = 10.13SLNSLTCTGYY1077 pKa = 10.11PNTSVGNPTITLAPGDD1093 pKa = 4.78DD1094 pKa = 4.04VTCTLVNDD1102 pKa = 5.56DD1103 pKa = 3.75ILIPVTVEE1111 pKa = 3.68KK1112 pKa = 11.02SDD1114 pKa = 3.61GTVEE1118 pKa = 3.92QLADD1122 pKa = 3.63GTWRR1126 pKa = 11.84ITYY1129 pKa = 9.13EE1130 pKa = 4.03VVVANGSPTLATRR1143 pKa = 11.84FSLTDD1148 pKa = 3.22TPQFDD1153 pKa = 3.6TSFTVLTQGWEE1164 pKa = 4.06GSPNVTDD1171 pKa = 3.83VPIEE1175 pKa = 4.2GGGVYY1180 pKa = 9.79TFTYY1184 pKa = 10.64VVTAEE1189 pKa = 4.11ANIEE1193 pKa = 4.02PVAPGALVCSPTNGGGFFNSATVTFPSGTDD1223 pKa = 3.04TDD1225 pKa = 3.95TGCAVPGAPTVQKK1238 pKa = 9.12TAQTAVQNPATGEE1251 pKa = 3.61WTLRR1255 pKa = 11.84YY1256 pKa = 9.19LVAVSNPTPIPLSYY1270 pKa = 10.07TLSDD1274 pKa = 3.35TAAPLPAGVAGGAWSASDD1292 pKa = 3.66PTPVGGGTFVRR1303 pKa = 11.84NAGWSGAGEE1312 pKa = 4.42LATGTLPAGAVHH1324 pKa = 7.0TYY1326 pKa = 10.58AVTRR1330 pKa = 11.84VVTVAASVSEE1340 pKa = 4.0AALTCGSTPADD1351 pKa = 3.95GGGVWNTASVTNGITVTEE1369 pKa = 4.38SSDD1372 pKa = 3.52CVDD1375 pKa = 3.24IEE1377 pKa = 4.36RR1378 pKa = 11.84PAVTIAKK1385 pKa = 8.14TVTDD1389 pKa = 3.87TRR1391 pKa = 11.84QLADD1395 pKa = 3.74GTWEE1399 pKa = 3.62IVYY1402 pKa = 10.44DD1403 pKa = 3.9VVVTNSSTTLAAVYY1417 pKa = 10.5SLADD1421 pKa = 3.46SLQFGGDD1428 pKa = 3.02ITVEE1432 pKa = 4.28GASWTGPTSGDD1443 pKa = 3.48FAPDD1447 pKa = 3.24GTAEE1451 pKa = 4.16LATDD1455 pKa = 3.9RR1456 pKa = 11.84VLAPLVGDD1464 pKa = 4.38AGVEE1468 pKa = 4.46TYY1470 pKa = 11.01VVTARR1475 pKa = 11.84ATVDD1479 pKa = 3.7LAAWEE1484 pKa = 4.79GDD1486 pKa = 3.66TLSCTDD1492 pKa = 4.54EE1493 pKa = 4.78DD1494 pKa = 4.73PPTAGGFLNVATVTVNGVGIPADD1517 pKa = 3.94DD1518 pKa = 4.18CSEE1521 pKa = 4.49PGLPTIAKK1529 pKa = 9.31QGVAATQDD1537 pKa = 3.54PSDD1540 pKa = 3.84PSAWTVTYY1548 pKa = 10.08EE1549 pKa = 4.04VTVTSGGFDD1558 pKa = 3.19TFYY1561 pKa = 11.31SLSDD1565 pKa = 3.51TPAFAAGIALNAGTAQRR1582 pKa = 11.84TDD1584 pKa = 3.1IADD1587 pKa = 3.67QPVLPISPGADD1598 pKa = 2.9FVTDD1602 pKa = 3.3VALAADD1608 pKa = 3.87ATHH1611 pKa = 6.7VYY1613 pKa = 9.74LVSWSVEE1620 pKa = 3.79VTDD1623 pKa = 6.14AFTEE1627 pKa = 4.27DD1628 pKa = 4.75DD1629 pKa = 4.21ADD1631 pKa = 4.0CTGEE1635 pKa = 4.08PGSGFFNSATLLVGDD1650 pKa = 4.68IPIDD1654 pKa = 3.94GSDD1657 pKa = 4.84CIPVADD1663 pKa = 4.1RR1664 pKa = 11.84VYY1666 pKa = 8.38PTIVKK1671 pKa = 8.67TVTGTDD1677 pKa = 3.43QDD1679 pKa = 4.23PSTGDD1684 pKa = 2.93WTITYY1689 pKa = 9.97AIEE1692 pKa = 3.9VTLAPVGPGNPDD1704 pKa = 2.99GLAAEE1709 pKa = 4.47YY1710 pKa = 11.04DD1711 pKa = 3.76LTDD1714 pKa = 3.48TLDD1717 pKa = 3.48FGGGIEE1723 pKa = 4.32VVEE1726 pKa = 4.34ATWAGQSSGTFDD1738 pKa = 4.04PVTDD1742 pKa = 4.05PAEE1745 pKa = 4.23LASDD1749 pKa = 3.59EE1750 pKa = 5.09AIAAGATHH1758 pKa = 6.94VYY1760 pKa = 10.18SVTVRR1765 pKa = 11.84ATVTAEE1771 pKa = 4.48AIDD1774 pKa = 4.57GGTTSCDD1781 pKa = 3.26PGEE1784 pKa = 4.37GQTGGFLNTALLSSGGEE1801 pKa = 4.02DD1802 pKa = 3.63TPVDD1806 pKa = 3.45ACAEE1810 pKa = 4.21PVFPTISKK1818 pKa = 10.58SSAQGGAVQQPDD1830 pKa = 4.26GSWALAYY1837 pKa = 9.9DD1838 pKa = 3.45ITVSYY1843 pKa = 10.03PSTDD1847 pKa = 3.34ADD1849 pKa = 3.55PRR1851 pKa = 11.84PSVGYY1856 pKa = 10.61VLTDD1860 pKa = 3.86VPDD1863 pKa = 4.06LPANVEE1869 pKa = 4.22LQGEE1873 pKa = 4.37WTATAGADD1881 pKa = 3.73TPAPDD1886 pKa = 5.16DD1887 pKa = 3.59PTFDD1891 pKa = 5.3GEE1893 pKa = 4.71GTWTIVTAALDD1904 pKa = 3.92PDD1906 pKa = 3.94EE1907 pKa = 6.62DD1908 pKa = 4.25GVTQHH1913 pKa = 6.23VYY1915 pKa = 10.28TITAVVRR1922 pKa = 11.84VTAPPVEE1929 pKa = 4.38PVAEE1933 pKa = 4.09CAEE1936 pKa = 4.19LEE1938 pKa = 4.18EE1939 pKa = 4.42TGIVVPNSATVTSGGYY1955 pKa = 7.58TADD1958 pKa = 4.84DD1959 pKa = 4.46DD1960 pKa = 4.27ACQVVQFDD1968 pKa = 4.08DD1969 pKa = 3.83VGVEE1973 pKa = 4.12KK1974 pKa = 10.88SADD1977 pKa = 3.48LAGEE1981 pKa = 4.21EE1982 pKa = 4.5TSVEE1986 pKa = 4.03PGDD1989 pKa = 3.51QFDD1992 pKa = 4.1YY1993 pKa = 11.35VITVTNTGTGPANEE2007 pKa = 4.28ILVFDD2012 pKa = 5.3DD2013 pKa = 4.42SLSEE2017 pKa = 3.84APYY2020 pKa = 9.6VGRR2023 pKa = 11.84VQLVAGTLSVEE2034 pKa = 4.3PAGLAYY2040 pKa = 9.49TDD2042 pKa = 4.66SSDD2045 pKa = 4.43LPGNVIDD2052 pKa = 6.39LEE2054 pKa = 4.16IDD2056 pKa = 3.25QLAVGEE2062 pKa = 4.43SATIRR2067 pKa = 11.84VTAEE2071 pKa = 3.61FLPAPVGGEE2080 pKa = 3.82PQVPPGEE2087 pKa = 4.92DD2088 pKa = 3.52PPAPTPPLDD2097 pKa = 4.75AIVNLVCVAAEE2108 pKa = 3.74FDD2110 pKa = 4.29GNPEE2114 pKa = 4.08NNCDD2118 pKa = 3.87DD2119 pKa = 4.71AEE2121 pKa = 4.38VPTRR2125 pKa = 11.84DD2126 pKa = 2.94ITASVYY2132 pKa = 8.46ATCQSDD2138 pKa = 3.87APLLGWVVAKK2148 pKa = 10.49SALLAGEE2155 pKa = 4.2PASLLWEE2162 pKa = 5.09LDD2164 pKa = 3.5PPSPGSVPPFVEE2176 pKa = 4.37FDD2178 pKa = 3.47EE2179 pKa = 4.9SDD2181 pKa = 3.35EE2182 pKa = 4.44GAVGPVWTGLADD2194 pKa = 3.32WPGAFFTPSGVAIDD2208 pKa = 4.02YY2209 pKa = 8.49PGWRR2213 pKa = 11.84PIEE2216 pKa = 4.26LSDD2219 pKa = 4.44VVPGSNPINYY2229 pKa = 7.99YY2230 pKa = 10.08IPGPEE2235 pKa = 4.41PRR2237 pKa = 11.84TEE2239 pKa = 3.58MTAADD2244 pKa = 4.62RR2245 pKa = 11.84LNLVYY2250 pKa = 10.76NGLILDD2256 pKa = 4.48DD2257 pKa = 5.05SEE2259 pKa = 6.32LDD2261 pKa = 3.86YY2262 pKa = 11.4AWRR2265 pKa = 11.84GASTVTLSVNPEE2277 pKa = 3.62LTFEE2281 pKa = 4.23VEE2283 pKa = 4.42YY2284 pKa = 10.46PPATPEE2290 pKa = 4.12CVQARR2295 pKa = 11.84HH2296 pKa = 5.0STVEE2300 pKa = 3.77IEE2302 pKa = 4.08KK2303 pKa = 8.62TASVEE2308 pKa = 4.02RR2309 pKa = 11.84TSAGQSFTYY2318 pKa = 10.17SLEE2321 pKa = 4.15VANVSDD2327 pKa = 4.49DD2328 pKa = 3.76SAAEE2332 pKa = 3.87AVVITDD2338 pKa = 5.83AIPADD2343 pKa = 3.83LRR2345 pKa = 11.84ITSVTWPGQGDD2356 pKa = 3.56AGVFPNWSSCAVTGQNAQGYY2376 pKa = 9.53GGVLNCTLFGPLQPVGANDD2395 pKa = 4.05SPSSAPTITLAATVNPASTAAVITNVAVVEE2425 pKa = 4.21YY2426 pKa = 8.68QTFGDD2431 pKa = 4.31PEE2433 pKa = 4.24DD2434 pKa = 4.09TGTDD2438 pKa = 3.39SDD2440 pKa = 4.5DD2441 pKa = 3.58ATVLLSGLPVTGGSPALPLIMLGFLALLGGVATLVVTRR2479 pKa = 11.84RR2480 pKa = 11.84RR2481 pKa = 11.84RR2482 pKa = 11.84KK2483 pKa = 9.1SATVDD2488 pKa = 3.4VQQ2490 pKa = 3.25
Molecular weight: 254.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6GE02|A0A4R6GE02_9MICO Deazaflavin-dependent oxidoreductase (Nitroreductase family) OS=Microbacterium sp. BK668 OX=2512118 GN=EV279_2538 PE=3 SV=1
MM1 pKa = 7.39RR2 pKa = 11.84RR3 pKa = 11.84SAIKK7 pKa = 10.26ALLRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84GSADD17 pKa = 3.3PSPASSSSARR27 pKa = 11.84LRR29 pKa = 11.84TAPLLPAGPRR39 pKa = 11.84VPQPRR44 pKa = 11.84TLRR47 pKa = 11.84QFSASPRR54 pKa = 11.84STQLPPTRR62 pKa = 11.84PQLPRR67 pKa = 11.84GPSFPLRR74 pKa = 11.84RR75 pKa = 11.84GMPRR79 pKa = 11.84PPLLPRR85 pKa = 11.84SARR88 pKa = 11.84RR89 pKa = 11.84SRR91 pKa = 11.84PPPRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84SSVRR104 pKa = 11.84PQPLCRR110 pKa = 11.84PAGLHH115 pKa = 5.55RR116 pKa = 11.84PRR118 pKa = 11.84RR119 pKa = 11.84PSSVFLRR126 pKa = 11.84RR127 pKa = 11.84PLHH130 pKa = 6.24RR131 pKa = 11.84VAPLPRR137 pKa = 11.84PRR139 pKa = 11.84EE140 pKa = 4.05PRR142 pKa = 11.84HH143 pKa = 5.44PRR145 pKa = 11.84PPFLRR150 pKa = 11.84RR151 pKa = 11.84RR152 pKa = 11.84IRR154 pKa = 11.84SARR157 pKa = 11.84HH158 pKa = 5.07RR159 pKa = 11.84VRR161 pKa = 11.84SAPPRR166 pKa = 11.84RR167 pKa = 11.84IRR169 pKa = 11.84PARR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84TGRR177 pKa = 11.84PDD179 pKa = 3.02SRR181 pKa = 11.84CSRR184 pKa = 11.84RR185 pKa = 11.84CRR187 pKa = 11.84RR188 pKa = 11.84PRR190 pKa = 11.84RR191 pKa = 11.84PPRR194 pKa = 11.84RR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84HH199 pKa = 5.37HH200 pKa = 6.57RR201 pKa = 11.84ALPPLRR207 pKa = 11.84APRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84RR213 pKa = 11.84RR214 pKa = 11.84PFPPGHH220 pKa = 6.12RR221 pKa = 11.84RR222 pKa = 11.84RR223 pKa = 11.84RR224 pKa = 11.84VTRR227 pKa = 11.84GLCSARR233 pKa = 11.84PAPRR237 pKa = 11.84PSSRR241 pKa = 11.84PRR243 pKa = 11.84DD244 pKa = 3.42RR245 pKa = 11.84MSTPRR250 pKa = 11.84SRR252 pKa = 11.84AGAHH256 pKa = 6.08PLRR259 pKa = 11.84SRR261 pKa = 11.84STQRR265 pKa = 11.84SRR267 pKa = 11.84ARR269 pKa = 11.84RR270 pKa = 11.84SRR272 pKa = 11.84SAQRR276 pKa = 11.84SSARR280 pKa = 11.84RR281 pKa = 11.84IRR283 pKa = 11.84ASSPARR289 pKa = 11.84QRR291 pKa = 11.84SPRR294 pKa = 11.84GTRR297 pKa = 11.84PLPLSSLRR305 pKa = 11.84PLPTSSARR313 pKa = 11.84PPSARR318 pKa = 11.84RR319 pKa = 11.84HH320 pKa = 4.39RR321 pKa = 11.84HH322 pKa = 4.28RR323 pKa = 11.84CRR325 pKa = 11.84CRR327 pKa = 11.84HH328 pKa = 4.48RR329 pKa = 11.84HH330 pKa = 5.34GPCSAWALLSGSPSLRR346 pKa = 11.84RR347 pKa = 11.84AAAFRR352 pKa = 11.84FSARR356 pKa = 11.84PRR358 pKa = 11.84STRR361 pKa = 11.84HH362 pKa = 5.5LIHH365 pKa = 6.75RR366 pKa = 11.84RR367 pKa = 11.84HH368 pKa = 6.69PGRR371 pKa = 11.84PSLRR375 pKa = 11.84RR376 pKa = 11.84PSPPSPARR384 pKa = 11.84PGLRR388 pKa = 11.84RR389 pKa = 11.84QPPGPPPRR397 pKa = 11.84RR398 pKa = 11.84MAASPATSS406 pKa = 2.94
MM1 pKa = 7.39RR2 pKa = 11.84RR3 pKa = 11.84SAIKK7 pKa = 10.26ALLRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84GSADD17 pKa = 3.3PSPASSSSARR27 pKa = 11.84LRR29 pKa = 11.84TAPLLPAGPRR39 pKa = 11.84VPQPRR44 pKa = 11.84TLRR47 pKa = 11.84QFSASPRR54 pKa = 11.84STQLPPTRR62 pKa = 11.84PQLPRR67 pKa = 11.84GPSFPLRR74 pKa = 11.84RR75 pKa = 11.84GMPRR79 pKa = 11.84PPLLPRR85 pKa = 11.84SARR88 pKa = 11.84RR89 pKa = 11.84SRR91 pKa = 11.84PPPRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84SSVRR104 pKa = 11.84PQPLCRR110 pKa = 11.84PAGLHH115 pKa = 5.55RR116 pKa = 11.84PRR118 pKa = 11.84RR119 pKa = 11.84PSSVFLRR126 pKa = 11.84RR127 pKa = 11.84PLHH130 pKa = 6.24RR131 pKa = 11.84VAPLPRR137 pKa = 11.84PRR139 pKa = 11.84EE140 pKa = 4.05PRR142 pKa = 11.84HH143 pKa = 5.44PRR145 pKa = 11.84PPFLRR150 pKa = 11.84RR151 pKa = 11.84RR152 pKa = 11.84IRR154 pKa = 11.84SARR157 pKa = 11.84HH158 pKa = 5.07RR159 pKa = 11.84VRR161 pKa = 11.84SAPPRR166 pKa = 11.84RR167 pKa = 11.84IRR169 pKa = 11.84PARR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84TGRR177 pKa = 11.84PDD179 pKa = 3.02SRR181 pKa = 11.84CSRR184 pKa = 11.84RR185 pKa = 11.84CRR187 pKa = 11.84RR188 pKa = 11.84PRR190 pKa = 11.84RR191 pKa = 11.84PPRR194 pKa = 11.84RR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84HH199 pKa = 5.37HH200 pKa = 6.57RR201 pKa = 11.84ALPPLRR207 pKa = 11.84APRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84RR213 pKa = 11.84RR214 pKa = 11.84PFPPGHH220 pKa = 6.12RR221 pKa = 11.84RR222 pKa = 11.84RR223 pKa = 11.84RR224 pKa = 11.84VTRR227 pKa = 11.84GLCSARR233 pKa = 11.84PAPRR237 pKa = 11.84PSSRR241 pKa = 11.84PRR243 pKa = 11.84DD244 pKa = 3.42RR245 pKa = 11.84MSTPRR250 pKa = 11.84SRR252 pKa = 11.84AGAHH256 pKa = 6.08PLRR259 pKa = 11.84SRR261 pKa = 11.84STQRR265 pKa = 11.84SRR267 pKa = 11.84ARR269 pKa = 11.84RR270 pKa = 11.84SRR272 pKa = 11.84SAQRR276 pKa = 11.84SSARR280 pKa = 11.84RR281 pKa = 11.84IRR283 pKa = 11.84ASSPARR289 pKa = 11.84QRR291 pKa = 11.84SPRR294 pKa = 11.84GTRR297 pKa = 11.84PLPLSSLRR305 pKa = 11.84PLPTSSARR313 pKa = 11.84PPSARR318 pKa = 11.84RR319 pKa = 11.84HH320 pKa = 4.39RR321 pKa = 11.84HH322 pKa = 4.28RR323 pKa = 11.84CRR325 pKa = 11.84CRR327 pKa = 11.84HH328 pKa = 4.48RR329 pKa = 11.84HH330 pKa = 5.34GPCSAWALLSGSPSLRR346 pKa = 11.84RR347 pKa = 11.84AAAFRR352 pKa = 11.84FSARR356 pKa = 11.84PRR358 pKa = 11.84STRR361 pKa = 11.84HH362 pKa = 5.5LIHH365 pKa = 6.75RR366 pKa = 11.84RR367 pKa = 11.84HH368 pKa = 6.69PGRR371 pKa = 11.84PSLRR375 pKa = 11.84RR376 pKa = 11.84PSPPSPARR384 pKa = 11.84PGLRR388 pKa = 11.84RR389 pKa = 11.84QPPGPPPRR397 pKa = 11.84RR398 pKa = 11.84MAASPATSS406 pKa = 2.94
Molecular weight: 46.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1141260 |
30 |
10586 |
343.9 |
36.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.714 ± 0.061 | 0.494 ± 0.009 |
6.366 ± 0.038 | 5.664 ± 0.049 |
3.13 ± 0.026 | 9.123 ± 0.054 |
1.898 ± 0.022 | 4.368 ± 0.031 |
1.748 ± 0.027 | 9.912 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.63 ± 0.018 | 1.841 ± 0.033 |
5.712 ± 0.036 | 2.674 ± 0.021 |
7.56 ± 0.065 | 5.618 ± 0.036 |
5.957 ± 0.061 | 8.999 ± 0.04 |
1.59 ± 0.02 | 2.0 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
