
Gammapapillomavirus 23
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
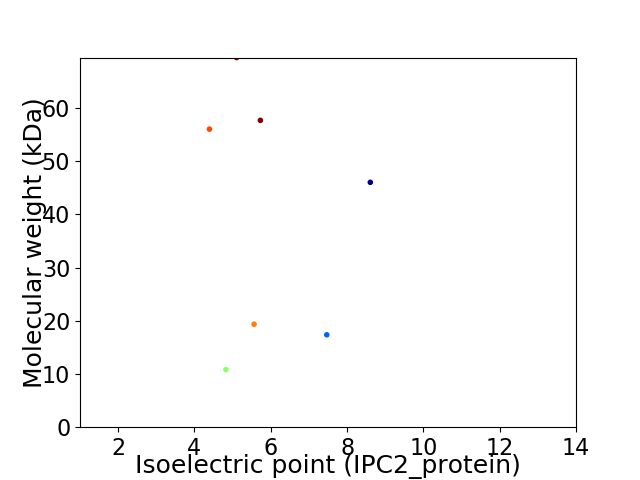
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
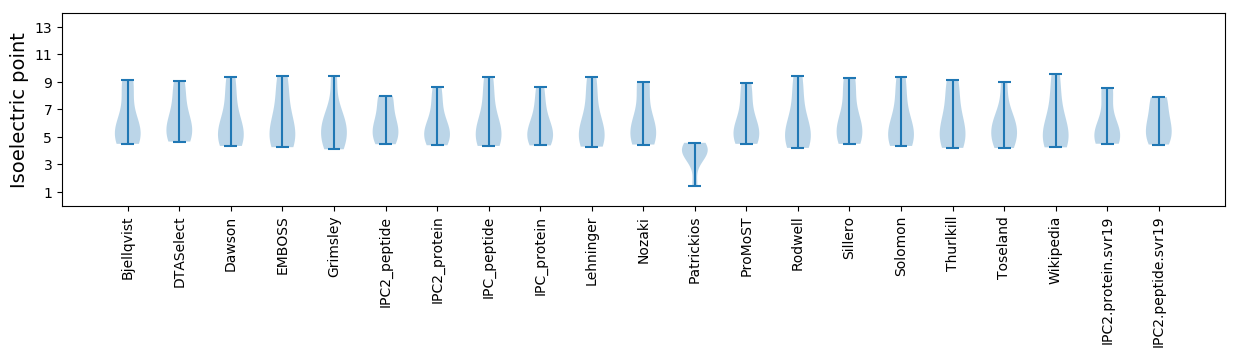
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2AM71|A0A2D2AM71_9PAPI Protein E7 OS=Gammapapillomavirus 23 OX=1961680 GN=E7 PE=3 SV=1
MM1 pKa = 7.45TSLKK5 pKa = 9.93RR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.55RR9 pKa = 11.84DD10 pKa = 3.47TVDD13 pKa = 2.81NLYY16 pKa = 10.32RR17 pKa = 11.84QCQLGADD24 pKa = 4.28CPPDD28 pKa = 3.29VRR30 pKa = 11.84NKK32 pKa = 10.25VEE34 pKa = 4.05GTTLADD40 pKa = 3.71KK41 pKa = 10.79LLQAFGSIIFLGGLGIGSGSGSGAVTAGRR70 pKa = 11.84TIPDD74 pKa = 4.56LAPDD78 pKa = 4.12ATPIRR83 pKa = 11.84PTRR86 pKa = 11.84PLRR89 pKa = 11.84PSSGPNTTRR98 pKa = 11.84PFSVPIDD105 pKa = 4.0RR106 pKa = 11.84IGVPGAGGRR115 pKa = 11.84PVTIDD120 pKa = 3.23ATSSSIVPLTEE131 pKa = 4.07AVPDD135 pKa = 3.96TVITLGDD142 pKa = 3.53PTIGVTTDD150 pKa = 2.68IAVDD154 pKa = 3.46VNPIEE159 pKa = 5.26LEE161 pKa = 4.58PINQGPDD168 pKa = 2.88TPAIINVTPIEE179 pKa = 4.35PPPVRR184 pKa = 11.84ILYY187 pKa = 10.48SDD189 pKa = 3.47TTTYY193 pKa = 11.18APPEE197 pKa = 4.08TVQIAFTDD205 pKa = 3.67PNLNVFVDD213 pKa = 4.25PASTGQHH220 pKa = 5.78IGLEE224 pKa = 4.38EE225 pKa = 4.72IEE227 pKa = 5.14LDD229 pKa = 4.06TIGGPSTFEE238 pKa = 3.8IEE240 pKa = 4.02EE241 pKa = 4.24AAPTTSTPLEE251 pKa = 4.14RR252 pKa = 11.84LGQVYY257 pKa = 10.23NRR259 pKa = 11.84ARR261 pKa = 11.84QYY263 pKa = 10.24YY264 pKa = 7.78NRR266 pKa = 11.84RR267 pKa = 11.84VQQVPTRR274 pKa = 11.84NIDD277 pKa = 3.57FLGQPSRR284 pKa = 11.84AVLFGFEE291 pKa = 4.34NPAFLDD297 pKa = 3.85DD298 pKa = 3.63VTLEE302 pKa = 4.05FEE304 pKa = 3.78QDD306 pKa = 3.37LRR308 pKa = 11.84EE309 pKa = 4.16VAAAPDD315 pKa = 3.44EE316 pKa = 4.38DD317 pKa = 4.16FRR319 pKa = 11.84DD320 pKa = 3.64VRR322 pKa = 11.84QLHH325 pKa = 6.26RR326 pKa = 11.84PTFSITDD333 pKa = 3.3EE334 pKa = 4.08GLIRR338 pKa = 11.84LSRR341 pKa = 11.84LGSRR345 pKa = 11.84ATMQTRR351 pKa = 11.84SGRR354 pKa = 11.84VFGSDD359 pKa = 2.15AHH361 pKa = 6.41FYY363 pKa = 10.68YY364 pKa = 10.54DD365 pKa = 3.26VSEE368 pKa = 4.21IPEE371 pKa = 4.17AGEE374 pKa = 4.92IEE376 pKa = 4.37MQDD379 pKa = 3.48LVTPGVPYY387 pKa = 10.24TIVDD391 pKa = 3.9PQAEE395 pKa = 4.37SSFVNALADD404 pKa = 3.48VTLFTEE410 pKa = 4.41DD411 pKa = 3.7QLVDD415 pKa = 4.23FYY417 pKa = 12.02NEE419 pKa = 4.2SFDD422 pKa = 3.73NAQLILEE429 pKa = 4.61GQTEE433 pKa = 4.16AEE435 pKa = 4.0EE436 pKa = 5.56RR437 pKa = 11.84YY438 pKa = 7.83TFPIPISTTAIKK450 pKa = 10.37PFVVDD455 pKa = 3.3IGNGIFYY462 pKa = 10.49SSPTQISTNNYY473 pKa = 8.06SPKK476 pKa = 10.08FPFIPVSPGVTINMYY491 pKa = 10.83SDD493 pKa = 5.37DD494 pKa = 4.42YY495 pKa = 11.78YY496 pKa = 11.61LDD498 pKa = 4.28PSLLKK503 pKa = 10.39RR504 pKa = 11.84KK505 pKa = 9.64RR506 pKa = 11.84KK507 pKa = 10.04RR508 pKa = 11.84SDD510 pKa = 3.14DD511 pKa = 3.61FF512 pKa = 6.03
MM1 pKa = 7.45TSLKK5 pKa = 9.93RR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.55RR9 pKa = 11.84DD10 pKa = 3.47TVDD13 pKa = 2.81NLYY16 pKa = 10.32RR17 pKa = 11.84QCQLGADD24 pKa = 4.28CPPDD28 pKa = 3.29VRR30 pKa = 11.84NKK32 pKa = 10.25VEE34 pKa = 4.05GTTLADD40 pKa = 3.71KK41 pKa = 10.79LLQAFGSIIFLGGLGIGSGSGSGAVTAGRR70 pKa = 11.84TIPDD74 pKa = 4.56LAPDD78 pKa = 4.12ATPIRR83 pKa = 11.84PTRR86 pKa = 11.84PLRR89 pKa = 11.84PSSGPNTTRR98 pKa = 11.84PFSVPIDD105 pKa = 4.0RR106 pKa = 11.84IGVPGAGGRR115 pKa = 11.84PVTIDD120 pKa = 3.23ATSSSIVPLTEE131 pKa = 4.07AVPDD135 pKa = 3.96TVITLGDD142 pKa = 3.53PTIGVTTDD150 pKa = 2.68IAVDD154 pKa = 3.46VNPIEE159 pKa = 5.26LEE161 pKa = 4.58PINQGPDD168 pKa = 2.88TPAIINVTPIEE179 pKa = 4.35PPPVRR184 pKa = 11.84ILYY187 pKa = 10.48SDD189 pKa = 3.47TTTYY193 pKa = 11.18APPEE197 pKa = 4.08TVQIAFTDD205 pKa = 3.67PNLNVFVDD213 pKa = 4.25PASTGQHH220 pKa = 5.78IGLEE224 pKa = 4.38EE225 pKa = 4.72IEE227 pKa = 5.14LDD229 pKa = 4.06TIGGPSTFEE238 pKa = 3.8IEE240 pKa = 4.02EE241 pKa = 4.24AAPTTSTPLEE251 pKa = 4.14RR252 pKa = 11.84LGQVYY257 pKa = 10.23NRR259 pKa = 11.84ARR261 pKa = 11.84QYY263 pKa = 10.24YY264 pKa = 7.78NRR266 pKa = 11.84RR267 pKa = 11.84VQQVPTRR274 pKa = 11.84NIDD277 pKa = 3.57FLGQPSRR284 pKa = 11.84AVLFGFEE291 pKa = 4.34NPAFLDD297 pKa = 3.85DD298 pKa = 3.63VTLEE302 pKa = 4.05FEE304 pKa = 3.78QDD306 pKa = 3.37LRR308 pKa = 11.84EE309 pKa = 4.16VAAAPDD315 pKa = 3.44EE316 pKa = 4.38DD317 pKa = 4.16FRR319 pKa = 11.84DD320 pKa = 3.64VRR322 pKa = 11.84QLHH325 pKa = 6.26RR326 pKa = 11.84PTFSITDD333 pKa = 3.3EE334 pKa = 4.08GLIRR338 pKa = 11.84LSRR341 pKa = 11.84LGSRR345 pKa = 11.84ATMQTRR351 pKa = 11.84SGRR354 pKa = 11.84VFGSDD359 pKa = 2.15AHH361 pKa = 6.41FYY363 pKa = 10.68YY364 pKa = 10.54DD365 pKa = 3.26VSEE368 pKa = 4.21IPEE371 pKa = 4.17AGEE374 pKa = 4.92IEE376 pKa = 4.37MQDD379 pKa = 3.48LVTPGVPYY387 pKa = 10.24TIVDD391 pKa = 3.9PQAEE395 pKa = 4.37SSFVNALADD404 pKa = 3.48VTLFTEE410 pKa = 4.41DD411 pKa = 3.7QLVDD415 pKa = 4.23FYY417 pKa = 12.02NEE419 pKa = 4.2SFDD422 pKa = 3.73NAQLILEE429 pKa = 4.61GQTEE433 pKa = 4.16AEE435 pKa = 4.0EE436 pKa = 5.56RR437 pKa = 11.84YY438 pKa = 7.83TFPIPISTTAIKK450 pKa = 10.37PFVVDD455 pKa = 3.3IGNGIFYY462 pKa = 10.49SSPTQISTNNYY473 pKa = 8.06SPKK476 pKa = 10.08FPFIPVSPGVTINMYY491 pKa = 10.83SDD493 pKa = 5.37DD494 pKa = 4.42YY495 pKa = 11.78YY496 pKa = 11.61LDD498 pKa = 4.28PSLLKK503 pKa = 10.39RR504 pKa = 11.84KK505 pKa = 9.64RR506 pKa = 11.84KK507 pKa = 10.04RR508 pKa = 11.84SDD510 pKa = 3.14DD511 pKa = 3.61FF512 pKa = 6.03
Molecular weight: 56.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2AM70|A0A2D2AM70_9PAPI Minor capsid protein L2 OS=Gammapapillomavirus 23 OX=1961680 GN=L2 PE=3 SV=1
MM1 pKa = 7.03NQADD5 pKa = 4.0LTSRR9 pKa = 11.84SDD11 pKa = 3.46ALQEE15 pKa = 3.93RR16 pKa = 11.84LMNLYY21 pKa = 10.43EE22 pKa = 4.63SGAKK26 pKa = 8.78TLEE29 pKa = 3.77AQIEE33 pKa = 4.15HH34 pKa = 5.96WQLVRR39 pKa = 11.84KK40 pKa = 10.11LNVLYY45 pKa = 10.7YY46 pKa = 8.8YY47 pKa = 10.76ARR49 pKa = 11.84KK50 pKa = 9.81EE51 pKa = 4.21KK52 pKa = 10.35FSHH55 pKa = 7.04LGLQPLPSLTVSEE68 pKa = 4.7YY69 pKa = 10.45KK70 pKa = 10.56SKK72 pKa = 10.8EE73 pKa = 3.76AIQIVLLLRR82 pKa = 11.84SLQNSPYY89 pKa = 10.83AEE91 pKa = 4.3EE92 pKa = 4.17EE93 pKa = 4.25WNLSDD98 pKa = 3.83TSAEE102 pKa = 4.53LIHH105 pKa = 6.36TPPKK109 pKa = 9.49NTFKK113 pKa = 10.76KK114 pKa = 9.68GPYY117 pKa = 9.47RR118 pKa = 11.84VDD120 pKa = 2.16VWFDD124 pKa = 3.65HH125 pKa = 6.58NPQNSFPYY133 pKa = 9.31TNYY136 pKa = 10.99DD137 pKa = 3.9FIYY140 pKa = 9.39YY141 pKa = 9.64QDD143 pKa = 4.78YY144 pKa = 11.12DD145 pKa = 4.26EE146 pKa = 4.82QWHH149 pKa = 5.54KK150 pKa = 9.39TAGLVDD156 pKa = 3.44INGFYY161 pKa = 10.96YY162 pKa = 10.39EE163 pKa = 4.76EE164 pKa = 4.72PNGDD168 pKa = 3.02RR169 pKa = 11.84VYY171 pKa = 11.39YY172 pKa = 10.1FLFEE176 pKa = 4.43GDD178 pKa = 3.05AARR181 pKa = 11.84YY182 pKa = 10.03GEE184 pKa = 4.22TGQWTVQFKK193 pKa = 9.07NTTLSTSIPSSHH205 pKa = 6.75RR206 pKa = 11.84PQSTISPQGSISSSSDD222 pKa = 3.34SISPTQSFNSKK233 pKa = 9.64HH234 pKa = 5.75SRR236 pKa = 11.84SHH238 pKa = 5.91EE239 pKa = 4.2SEE241 pKa = 4.05EE242 pKa = 4.62GNASSSTGTPPQTPIRR258 pKa = 11.84QRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84EE265 pKa = 4.02GEE267 pKa = 3.9PTSTTGEE274 pKa = 3.84GSRR277 pKa = 11.84AKK279 pKa = 10.06RR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84GGHH285 pKa = 4.73TPVVRR290 pKa = 11.84AGVSAAEE297 pKa = 3.99VGSRR301 pKa = 11.84HH302 pKa = 6.07RR303 pKa = 11.84GVPTSGLTRR312 pKa = 11.84LARR315 pKa = 11.84LEE317 pKa = 4.14AEE319 pKa = 4.35ARR321 pKa = 11.84DD322 pKa = 3.73PLILIFKK329 pKa = 9.89GRR331 pKa = 11.84SNQLKK336 pKa = 8.03CWRR339 pKa = 11.84YY340 pKa = 9.77RR341 pKa = 11.84LPKK344 pKa = 10.45DD345 pKa = 3.46SFNVATTVFRR355 pKa = 11.84WAGEE359 pKa = 4.19EE360 pKa = 4.02EE361 pKa = 4.19DD362 pKa = 5.68DD363 pKa = 4.75SYY365 pKa = 11.78LSHH368 pKa = 7.28RR369 pKa = 11.84MLVAFNNQTQRR380 pKa = 11.84KK381 pKa = 8.19YY382 pKa = 10.12FLKK385 pKa = 10.47SVSIPRR391 pKa = 11.84GVSFAYY397 pKa = 10.13GQLDD401 pKa = 3.52SLL403 pKa = 4.56
MM1 pKa = 7.03NQADD5 pKa = 4.0LTSRR9 pKa = 11.84SDD11 pKa = 3.46ALQEE15 pKa = 3.93RR16 pKa = 11.84LMNLYY21 pKa = 10.43EE22 pKa = 4.63SGAKK26 pKa = 8.78TLEE29 pKa = 3.77AQIEE33 pKa = 4.15HH34 pKa = 5.96WQLVRR39 pKa = 11.84KK40 pKa = 10.11LNVLYY45 pKa = 10.7YY46 pKa = 8.8YY47 pKa = 10.76ARR49 pKa = 11.84KK50 pKa = 9.81EE51 pKa = 4.21KK52 pKa = 10.35FSHH55 pKa = 7.04LGLQPLPSLTVSEE68 pKa = 4.7YY69 pKa = 10.45KK70 pKa = 10.56SKK72 pKa = 10.8EE73 pKa = 3.76AIQIVLLLRR82 pKa = 11.84SLQNSPYY89 pKa = 10.83AEE91 pKa = 4.3EE92 pKa = 4.17EE93 pKa = 4.25WNLSDD98 pKa = 3.83TSAEE102 pKa = 4.53LIHH105 pKa = 6.36TPPKK109 pKa = 9.49NTFKK113 pKa = 10.76KK114 pKa = 9.68GPYY117 pKa = 9.47RR118 pKa = 11.84VDD120 pKa = 2.16VWFDD124 pKa = 3.65HH125 pKa = 6.58NPQNSFPYY133 pKa = 9.31TNYY136 pKa = 10.99DD137 pKa = 3.9FIYY140 pKa = 9.39YY141 pKa = 9.64QDD143 pKa = 4.78YY144 pKa = 11.12DD145 pKa = 4.26EE146 pKa = 4.82QWHH149 pKa = 5.54KK150 pKa = 9.39TAGLVDD156 pKa = 3.44INGFYY161 pKa = 10.96YY162 pKa = 10.39EE163 pKa = 4.76EE164 pKa = 4.72PNGDD168 pKa = 3.02RR169 pKa = 11.84VYY171 pKa = 11.39YY172 pKa = 10.1FLFEE176 pKa = 4.43GDD178 pKa = 3.05AARR181 pKa = 11.84YY182 pKa = 10.03GEE184 pKa = 4.22TGQWTVQFKK193 pKa = 9.07NTTLSTSIPSSHH205 pKa = 6.75RR206 pKa = 11.84PQSTISPQGSISSSSDD222 pKa = 3.34SISPTQSFNSKK233 pKa = 9.64HH234 pKa = 5.75SRR236 pKa = 11.84SHH238 pKa = 5.91EE239 pKa = 4.2SEE241 pKa = 4.05EE242 pKa = 4.62GNASSSTGTPPQTPIRR258 pKa = 11.84QRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84EE265 pKa = 4.02GEE267 pKa = 3.9PTSTTGEE274 pKa = 3.84GSRR277 pKa = 11.84AKK279 pKa = 10.06RR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84GGHH285 pKa = 4.73TPVVRR290 pKa = 11.84AGVSAAEE297 pKa = 3.99VGSRR301 pKa = 11.84HH302 pKa = 6.07RR303 pKa = 11.84GVPTSGLTRR312 pKa = 11.84LARR315 pKa = 11.84LEE317 pKa = 4.14AEE319 pKa = 4.35ARR321 pKa = 11.84DD322 pKa = 3.73PLILIFKK329 pKa = 9.89GRR331 pKa = 11.84SNQLKK336 pKa = 8.03CWRR339 pKa = 11.84YY340 pKa = 9.77RR341 pKa = 11.84LPKK344 pKa = 10.45DD345 pKa = 3.46SFNVATTVFRR355 pKa = 11.84WAGEE359 pKa = 4.19EE360 pKa = 4.02EE361 pKa = 4.19DD362 pKa = 5.68DD363 pKa = 4.75SYY365 pKa = 11.78LSHH368 pKa = 7.28RR369 pKa = 11.84MLVAFNNQTQRR380 pKa = 11.84KK381 pKa = 8.19YY382 pKa = 10.12FLKK385 pKa = 10.47SVSIPRR391 pKa = 11.84GVSFAYY397 pKa = 10.13GQLDD401 pKa = 3.52SLL403 pKa = 4.56
Molecular weight: 46.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2433 |
97 |
604 |
347.6 |
39.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.425 ± 0.358 | 2.261 ± 0.829 |
6.535 ± 0.528 | 6.494 ± 0.296 |
4.809 ± 0.551 | 5.302 ± 0.767 |
1.644 ± 0.302 | 5.179 ± 0.593 |
5.014 ± 0.889 | 9.042 ± 0.601 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.808 ± 0.348 | 5.302 ± 0.567 |
6.247 ± 1.053 | 4.316 ± 0.414 |
6.042 ± 0.671 | 6.987 ± 0.819 |
6.864 ± 0.598 | 5.59 ± 0.443 |
1.315 ± 0.34 | 3.822 ± 0.421 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
