
Tomato yellow mottle-associated virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Tomato yellow mottle-associated cytorhabdovirus
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
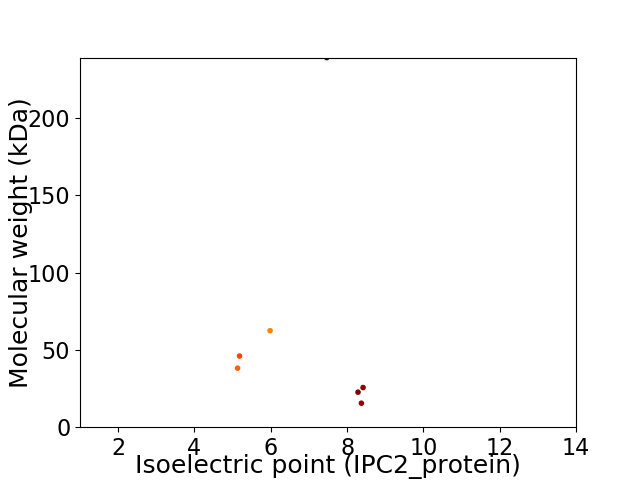
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U9W1C2|A0A1U9W1C2_9RHAB p3 protein OS=Tomato yellow mottle-associated virus OX=1967841 GN=P3 PE=4 SV=1
MM1 pKa = 6.91NTLALQLSNTDD12 pKa = 3.45YY13 pKa = 11.8DD14 pKa = 4.11DD15 pKa = 4.37LASITVAPGGSNVAWNDD32 pKa = 2.92EE33 pKa = 4.24DD34 pKa = 3.73VLSIRR39 pKa = 11.84RR40 pKa = 11.84YY41 pKa = 9.59SLAVMDD47 pKa = 4.56SPTMVLHH54 pKa = 6.41GGYY57 pKa = 10.14VFEE60 pKa = 4.81SLNNNARR67 pKa = 11.84VDD69 pKa = 4.12TNVLMSTVHH78 pKa = 6.5LAANLRR84 pKa = 11.84DD85 pKa = 3.89PDD87 pKa = 5.21NITQCLLTTPPHH99 pKa = 6.05SRR101 pKa = 11.84PAIIQTTLPTIRR113 pKa = 11.84AIQEE117 pKa = 4.51DD118 pKa = 3.71ITEE121 pKa = 4.23EE122 pKa = 4.02EE123 pKa = 4.7RR124 pKa = 11.84NTLALLNVDD133 pKa = 4.21VNIEE137 pKa = 4.03NQDD140 pKa = 3.52NGNQNQQGEE149 pKa = 4.64NVQVDD154 pKa = 3.52QGSIDD159 pKa = 3.73MKK161 pKa = 10.31AAAYY165 pKa = 9.92AYY167 pKa = 10.06VCAFLMRR174 pKa = 11.84LQCRR178 pKa = 11.84SAQNVSGGLRR188 pKa = 11.84RR189 pKa = 11.84AIDD192 pKa = 3.97RR193 pKa = 11.84FKK195 pKa = 10.17TWYY198 pKa = 10.13DD199 pKa = 3.0GSTDD203 pKa = 3.15IFDD206 pKa = 5.09EE207 pKa = 4.32ISFSIDD213 pKa = 2.83ALNQLKK219 pKa = 9.93EE220 pKa = 4.34AISRR224 pKa = 11.84KK225 pKa = 9.77PEE227 pKa = 3.71ITSTWVLYY235 pKa = 10.64LATTEE240 pKa = 4.19NEE242 pKa = 3.91KK243 pKa = 9.07TLLKK247 pKa = 10.15QSKK250 pKa = 10.41GMIEE254 pKa = 3.86YY255 pKa = 10.64LGLQVFSYY263 pKa = 10.79QGMHH267 pKa = 7.52ALTQVLALHH276 pKa = 5.89QMSKK280 pKa = 10.69VPLRR284 pKa = 11.84DD285 pKa = 3.19LMMEE289 pKa = 4.58LDD291 pKa = 4.03SPLTRR296 pKa = 11.84DD297 pKa = 3.49GLRR300 pKa = 11.84EE301 pKa = 3.48ISNILKK307 pKa = 9.59NHH309 pKa = 5.6EE310 pKa = 4.19RR311 pKa = 11.84TNRR314 pKa = 11.84APEE317 pKa = 3.79RR318 pKa = 11.84KK319 pKa = 7.95TYY321 pKa = 10.39FRR323 pKa = 11.84YY324 pKa = 10.21SRR326 pKa = 11.84VWDD329 pKa = 3.63TKK331 pKa = 11.22YY332 pKa = 10.27FAQLQSKK339 pKa = 7.42TCVPLLYY346 pKa = 10.2VAAVAVRR353 pKa = 11.84DD354 pKa = 3.75ISANTTSDD362 pKa = 3.05PTQIYY367 pKa = 9.94ALQNIGSAMKK377 pKa = 10.55ASLDD381 pKa = 3.44RR382 pKa = 11.84VAAALTSYY390 pKa = 10.88LVDD393 pKa = 3.49KK394 pKa = 10.54SYY396 pKa = 11.63RR397 pKa = 11.84DD398 pKa = 3.63AKK400 pKa = 10.76SGSVWDD406 pKa = 3.8AATPTEE412 pKa = 4.09
MM1 pKa = 6.91NTLALQLSNTDD12 pKa = 3.45YY13 pKa = 11.8DD14 pKa = 4.11DD15 pKa = 4.37LASITVAPGGSNVAWNDD32 pKa = 2.92EE33 pKa = 4.24DD34 pKa = 3.73VLSIRR39 pKa = 11.84RR40 pKa = 11.84YY41 pKa = 9.59SLAVMDD47 pKa = 4.56SPTMVLHH54 pKa = 6.41GGYY57 pKa = 10.14VFEE60 pKa = 4.81SLNNNARR67 pKa = 11.84VDD69 pKa = 4.12TNVLMSTVHH78 pKa = 6.5LAANLRR84 pKa = 11.84DD85 pKa = 3.89PDD87 pKa = 5.21NITQCLLTTPPHH99 pKa = 6.05SRR101 pKa = 11.84PAIIQTTLPTIRR113 pKa = 11.84AIQEE117 pKa = 4.51DD118 pKa = 3.71ITEE121 pKa = 4.23EE122 pKa = 4.02EE123 pKa = 4.7RR124 pKa = 11.84NTLALLNVDD133 pKa = 4.21VNIEE137 pKa = 4.03NQDD140 pKa = 3.52NGNQNQQGEE149 pKa = 4.64NVQVDD154 pKa = 3.52QGSIDD159 pKa = 3.73MKK161 pKa = 10.31AAAYY165 pKa = 9.92AYY167 pKa = 10.06VCAFLMRR174 pKa = 11.84LQCRR178 pKa = 11.84SAQNVSGGLRR188 pKa = 11.84RR189 pKa = 11.84AIDD192 pKa = 3.97RR193 pKa = 11.84FKK195 pKa = 10.17TWYY198 pKa = 10.13DD199 pKa = 3.0GSTDD203 pKa = 3.15IFDD206 pKa = 5.09EE207 pKa = 4.32ISFSIDD213 pKa = 2.83ALNQLKK219 pKa = 9.93EE220 pKa = 4.34AISRR224 pKa = 11.84KK225 pKa = 9.77PEE227 pKa = 3.71ITSTWVLYY235 pKa = 10.64LATTEE240 pKa = 4.19NEE242 pKa = 3.91KK243 pKa = 9.07TLLKK247 pKa = 10.15QSKK250 pKa = 10.41GMIEE254 pKa = 3.86YY255 pKa = 10.64LGLQVFSYY263 pKa = 10.79QGMHH267 pKa = 7.52ALTQVLALHH276 pKa = 5.89QMSKK280 pKa = 10.69VPLRR284 pKa = 11.84DD285 pKa = 3.19LMMEE289 pKa = 4.58LDD291 pKa = 4.03SPLTRR296 pKa = 11.84DD297 pKa = 3.49GLRR300 pKa = 11.84EE301 pKa = 3.48ISNILKK307 pKa = 9.59NHH309 pKa = 5.6EE310 pKa = 4.19RR311 pKa = 11.84TNRR314 pKa = 11.84APEE317 pKa = 3.79RR318 pKa = 11.84KK319 pKa = 7.95TYY321 pKa = 10.39FRR323 pKa = 11.84YY324 pKa = 10.21SRR326 pKa = 11.84VWDD329 pKa = 3.63TKK331 pKa = 11.22YY332 pKa = 10.27FAQLQSKK339 pKa = 7.42TCVPLLYY346 pKa = 10.2VAAVAVRR353 pKa = 11.84DD354 pKa = 3.75ISANTTSDD362 pKa = 3.05PTQIYY367 pKa = 9.94ALQNIGSAMKK377 pKa = 10.55ASLDD381 pKa = 3.44RR382 pKa = 11.84VAAALTSYY390 pKa = 10.88LVDD393 pKa = 3.49KK394 pKa = 10.54SYY396 pKa = 11.63RR397 pKa = 11.84DD398 pKa = 3.63AKK400 pKa = 10.76SGSVWDD406 pKa = 3.8AATPTEE412 pKa = 4.09
Molecular weight: 46.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U9W1C3|A0A1U9W1C3_9RHAB M protein OS=Tomato yellow mottle-associated virus OX=1967841 GN=M PE=4 SV=1
MM1 pKa = 7.72ASVFKK6 pKa = 10.82FNVKK10 pKa = 9.31TEE12 pKa = 4.11LTMSEE17 pKa = 3.92EE18 pKa = 4.42DD19 pKa = 3.83GVVSLTKK26 pKa = 10.4RR27 pKa = 11.84LNLWQTLKK35 pKa = 10.95SKK37 pKa = 9.76WMEE40 pKa = 3.69NVQLSNINFSFEE52 pKa = 4.24SRR54 pKa = 11.84GGPLAEE60 pKa = 4.09GRR62 pKa = 11.84VTASVHH68 pKa = 6.28DD69 pKa = 4.01NRR71 pKa = 11.84ISDD74 pKa = 3.93DD75 pKa = 3.74TSDD78 pKa = 3.6NTLKK82 pKa = 10.89SVTFSVTQDD91 pKa = 2.92ISFSWNYY98 pKa = 9.84NVNFEE103 pKa = 4.21VDD105 pKa = 3.48SLRR108 pKa = 11.84HH109 pKa = 5.2EE110 pKa = 4.37RR111 pKa = 11.84EE112 pKa = 3.87SPLILVTKK120 pKa = 10.58ISEE123 pKa = 4.2CNMRR127 pKa = 11.84PGYY130 pKa = 10.35SLGQIRR136 pKa = 11.84VQIEE140 pKa = 4.02MTTSDD145 pKa = 2.95KK146 pKa = 10.18MIRR149 pKa = 11.84RR150 pKa = 11.84IAKK153 pKa = 9.77SPVIKK158 pKa = 10.59LFRR161 pKa = 11.84DD162 pKa = 3.26HH163 pKa = 7.25KK164 pKa = 9.29MSEE167 pKa = 3.85NEE169 pKa = 3.65RR170 pKa = 11.84LGRR173 pKa = 11.84RR174 pKa = 11.84TSIEE178 pKa = 3.93RR179 pKa = 11.84LEE181 pKa = 4.25AAQNKK186 pKa = 9.03PLTIKK191 pKa = 10.35EE192 pKa = 4.58GPQGPYY198 pKa = 9.25MEE200 pKa = 5.89LDD202 pKa = 3.22RR203 pKa = 11.84SNSVMNIPRR212 pKa = 11.84TKK214 pKa = 10.48YY215 pKa = 10.26YY216 pKa = 10.09VAAPAKK222 pKa = 9.93EE223 pKa = 3.9II224 pKa = 3.95
MM1 pKa = 7.72ASVFKK6 pKa = 10.82FNVKK10 pKa = 9.31TEE12 pKa = 4.11LTMSEE17 pKa = 3.92EE18 pKa = 4.42DD19 pKa = 3.83GVVSLTKK26 pKa = 10.4RR27 pKa = 11.84LNLWQTLKK35 pKa = 10.95SKK37 pKa = 9.76WMEE40 pKa = 3.69NVQLSNINFSFEE52 pKa = 4.24SRR54 pKa = 11.84GGPLAEE60 pKa = 4.09GRR62 pKa = 11.84VTASVHH68 pKa = 6.28DD69 pKa = 4.01NRR71 pKa = 11.84ISDD74 pKa = 3.93DD75 pKa = 3.74TSDD78 pKa = 3.6NTLKK82 pKa = 10.89SVTFSVTQDD91 pKa = 2.92ISFSWNYY98 pKa = 9.84NVNFEE103 pKa = 4.21VDD105 pKa = 3.48SLRR108 pKa = 11.84HH109 pKa = 5.2EE110 pKa = 4.37RR111 pKa = 11.84EE112 pKa = 3.87SPLILVTKK120 pKa = 10.58ISEE123 pKa = 4.2CNMRR127 pKa = 11.84PGYY130 pKa = 10.35SLGQIRR136 pKa = 11.84VQIEE140 pKa = 4.02MTTSDD145 pKa = 2.95KK146 pKa = 10.18MIRR149 pKa = 11.84RR150 pKa = 11.84IAKK153 pKa = 9.77SPVIKK158 pKa = 10.59LFRR161 pKa = 11.84DD162 pKa = 3.26HH163 pKa = 7.25KK164 pKa = 9.29MSEE167 pKa = 3.85NEE169 pKa = 3.65RR170 pKa = 11.84LGRR173 pKa = 11.84RR174 pKa = 11.84TSIEE178 pKa = 3.93RR179 pKa = 11.84LEE181 pKa = 4.25AAQNKK186 pKa = 9.03PLTIKK191 pKa = 10.35EE192 pKa = 4.58GPQGPYY198 pKa = 9.25MEE200 pKa = 5.89LDD202 pKa = 3.22RR203 pKa = 11.84SNSVMNIPRR212 pKa = 11.84TKK214 pKa = 10.48YY215 pKa = 10.26YY216 pKa = 10.09VAAPAKK222 pKa = 9.93EE223 pKa = 3.9II224 pKa = 3.95
Molecular weight: 25.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3956 |
135 |
2085 |
565.1 |
64.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.207 ± 0.56 | 1.972 ± 0.439 |
6.143 ± 0.186 | 6.117 ± 0.385 |
2.932 ± 0.177 | 4.778 ± 0.408 |
2.275 ± 0.348 | 6.8 ± 0.489 |
6.143 ± 0.356 | 8.67 ± 0.934 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.564 ± 0.164 | 4.398 ± 0.461 |
4.019 ± 0.12 | 2.907 ± 0.526 |
6.218 ± 0.418 | 9.783 ± 0.585 |
6.269 ± 0.572 | 6.446 ± 0.443 |
1.542 ± 0.269 | 3.817 ± 0.461 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
