
Sewage-associated circular DNA virus-29
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
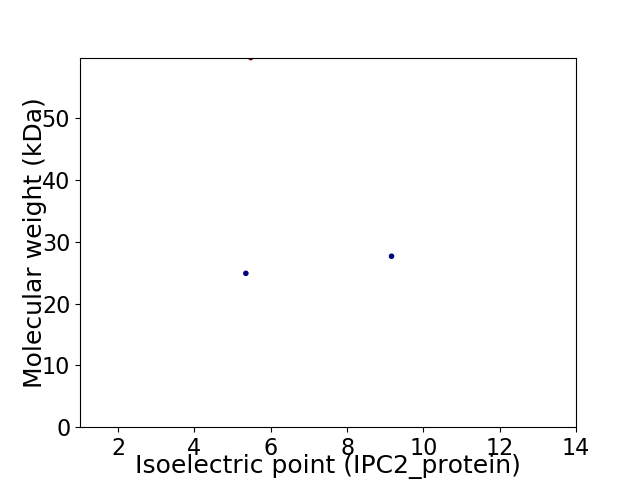
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGN4|A0A0B4UGN4_9VIRU Coat protein OS=Sewage-associated circular DNA virus-29 OX=1592096 PE=4 SV=1
MM1 pKa = 7.46FNYY4 pKa = 9.74QRR6 pKa = 11.84FQQDD10 pKa = 2.58EE11 pKa = 4.61RR12 pKa = 11.84EE13 pKa = 4.31LKK15 pKa = 10.35FCYY18 pKa = 9.85TNIGVSSLSWVGQRR32 pKa = 11.84FPVNRR37 pKa = 11.84IPRR40 pKa = 11.84GYY42 pKa = 10.95DD43 pKa = 2.89NVNQRR48 pKa = 11.84TGRR51 pKa = 11.84VITQHH56 pKa = 4.96FCKK59 pKa = 10.44LRR61 pKa = 11.84GFLQLQKK68 pKa = 9.91TEE70 pKa = 5.08DD71 pKa = 3.91IPWTGPNGDD80 pKa = 4.71FSPSQITRR88 pKa = 11.84LMIIEE93 pKa = 4.75DD94 pKa = 4.19LQPTGAGFTLTDD106 pKa = 4.73LFQEE110 pKa = 4.77TPSQYY115 pKa = 11.51ALPSDD120 pKa = 4.3FKK122 pKa = 11.0FSNINRR128 pKa = 11.84FRR130 pKa = 11.84IWFDD134 pKa = 2.7KK135 pKa = 10.78SYY137 pKa = 10.71PFDD140 pKa = 3.61AFAYY144 pKa = 6.45KK145 pKa = 10.06TGVIEE150 pKa = 4.37GFVNRR155 pKa = 11.84TIYY158 pKa = 10.22WIDD161 pKa = 3.42EE162 pKa = 4.76DD163 pKa = 4.25IDD165 pKa = 3.7LHH167 pKa = 6.61GLQTVYY173 pKa = 10.6GASPGVDD180 pKa = 2.74ILTSSLTFVAVSTANTPPVGGTQNGTSGSFRR211 pKa = 11.84CTIGFYY217 pKa = 10.99DD218 pKa = 4.37DD219 pKa = 3.55
MM1 pKa = 7.46FNYY4 pKa = 9.74QRR6 pKa = 11.84FQQDD10 pKa = 2.58EE11 pKa = 4.61RR12 pKa = 11.84EE13 pKa = 4.31LKK15 pKa = 10.35FCYY18 pKa = 9.85TNIGVSSLSWVGQRR32 pKa = 11.84FPVNRR37 pKa = 11.84IPRR40 pKa = 11.84GYY42 pKa = 10.95DD43 pKa = 2.89NVNQRR48 pKa = 11.84TGRR51 pKa = 11.84VITQHH56 pKa = 4.96FCKK59 pKa = 10.44LRR61 pKa = 11.84GFLQLQKK68 pKa = 9.91TEE70 pKa = 5.08DD71 pKa = 3.91IPWTGPNGDD80 pKa = 4.71FSPSQITRR88 pKa = 11.84LMIIEE93 pKa = 4.75DD94 pKa = 4.19LQPTGAGFTLTDD106 pKa = 4.73LFQEE110 pKa = 4.77TPSQYY115 pKa = 11.51ALPSDD120 pKa = 4.3FKK122 pKa = 11.0FSNINRR128 pKa = 11.84FRR130 pKa = 11.84IWFDD134 pKa = 2.7KK135 pKa = 10.78SYY137 pKa = 10.71PFDD140 pKa = 3.61AFAYY144 pKa = 6.45KK145 pKa = 10.06TGVIEE150 pKa = 4.37GFVNRR155 pKa = 11.84TIYY158 pKa = 10.22WIDD161 pKa = 3.42EE162 pKa = 4.76DD163 pKa = 4.25IDD165 pKa = 3.7LHH167 pKa = 6.61GLQTVYY173 pKa = 10.6GASPGVDD180 pKa = 2.74ILTSSLTFVAVSTANTPPVGGTQNGTSGSFRR211 pKa = 11.84CTIGFYY217 pKa = 10.99DD218 pKa = 4.37DD219 pKa = 3.55
Molecular weight: 24.9 kDa
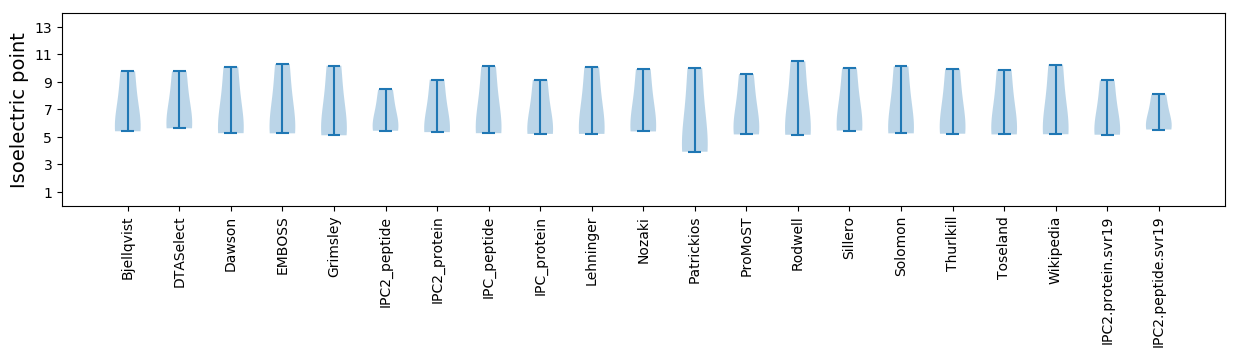
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGZ5|A0A0B4UGZ5_9VIRU ATP-dependent helicase Rep OS=Sewage-associated circular DNA virus-29 OX=1592096 PE=3 SV=1
MM1 pKa = 7.26SRR3 pKa = 11.84QVGQKK8 pKa = 8.19WKK10 pKa = 10.7APKK13 pKa = 9.81QLQAPRR19 pKa = 11.84NAFTNNNRR27 pKa = 11.84NPKK30 pKa = 10.17ANGRR34 pKa = 11.84SAPLATRR41 pKa = 11.84GFGDD45 pKa = 3.36MGILKK50 pKa = 9.9NRR52 pKa = 11.84EE53 pKa = 3.66KK54 pKa = 11.0KK55 pKa = 10.68FFDD58 pKa = 3.79RR59 pKa = 11.84STLGAPTGVSTTPVAYY75 pKa = 10.49LLHH78 pKa = 6.67NPAQGADD85 pKa = 3.32YY86 pKa = 10.17NQRR89 pKa = 11.84IGRR92 pKa = 11.84KK93 pKa = 6.49TVVKK97 pKa = 10.13SIYY100 pKa = 9.14IRR102 pKa = 11.84GRR104 pKa = 11.84MFVEE108 pKa = 4.34PVGLTPLPADD118 pKa = 3.25QWCPGQQARR127 pKa = 11.84LIVFIDD133 pKa = 3.66YY134 pKa = 10.33QPNGATPALTDD145 pKa = 4.56LLNSADD151 pKa = 4.18PASQLNADD159 pKa = 3.16NRR161 pKa = 11.84DD162 pKa = 3.24RR163 pKa = 11.84FKK165 pKa = 11.22VVKK168 pKa = 10.57DD169 pKa = 3.36KK170 pKa = 11.51VFTFGPVCVSNSDD183 pKa = 3.75LVSSEE188 pKa = 4.7RR189 pKa = 11.84SYY191 pKa = 11.92QDD193 pKa = 2.88FKK195 pKa = 11.42FFKK198 pKa = 10.29KK199 pKa = 10.72LNLEE203 pKa = 4.7CINNSTSTNAIADD216 pKa = 3.88IQSGALFAYY225 pKa = 9.63FIGTNVAGSTDD236 pKa = 4.18LVSVWTSRR244 pKa = 11.84TRR246 pKa = 11.84YY247 pKa = 10.1EE248 pKa = 4.66DD249 pKa = 3.16MM250 pKa = 5.39
MM1 pKa = 7.26SRR3 pKa = 11.84QVGQKK8 pKa = 8.19WKK10 pKa = 10.7APKK13 pKa = 9.81QLQAPRR19 pKa = 11.84NAFTNNNRR27 pKa = 11.84NPKK30 pKa = 10.17ANGRR34 pKa = 11.84SAPLATRR41 pKa = 11.84GFGDD45 pKa = 3.36MGILKK50 pKa = 9.9NRR52 pKa = 11.84EE53 pKa = 3.66KK54 pKa = 11.0KK55 pKa = 10.68FFDD58 pKa = 3.79RR59 pKa = 11.84STLGAPTGVSTTPVAYY75 pKa = 10.49LLHH78 pKa = 6.67NPAQGADD85 pKa = 3.32YY86 pKa = 10.17NQRR89 pKa = 11.84IGRR92 pKa = 11.84KK93 pKa = 6.49TVVKK97 pKa = 10.13SIYY100 pKa = 9.14IRR102 pKa = 11.84GRR104 pKa = 11.84MFVEE108 pKa = 4.34PVGLTPLPADD118 pKa = 3.25QWCPGQQARR127 pKa = 11.84LIVFIDD133 pKa = 3.66YY134 pKa = 10.33QPNGATPALTDD145 pKa = 4.56LLNSADD151 pKa = 4.18PASQLNADD159 pKa = 3.16NRR161 pKa = 11.84DD162 pKa = 3.24RR163 pKa = 11.84FKK165 pKa = 11.22VVKK168 pKa = 10.57DD169 pKa = 3.36KK170 pKa = 11.51VFTFGPVCVSNSDD183 pKa = 3.75LVSSEE188 pKa = 4.7RR189 pKa = 11.84SYY191 pKa = 11.92QDD193 pKa = 2.88FKK195 pKa = 11.42FFKK198 pKa = 10.29KK199 pKa = 10.72LNLEE203 pKa = 4.7CINNSTSTNAIADD216 pKa = 3.88IQSGALFAYY225 pKa = 9.63FIGTNVAGSTDD236 pKa = 4.18LVSVWTSRR244 pKa = 11.84TRR246 pKa = 11.84YY247 pKa = 10.1EE248 pKa = 4.66DD249 pKa = 3.16MM250 pKa = 5.39
Molecular weight: 27.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
996 |
219 |
527 |
332.0 |
37.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.827 ± 1.085 | 1.305 ± 0.033 |
7.129 ± 0.427 | 5.622 ± 1.714 |
4.217 ± 1.705 | 6.024 ± 1.004 |
1.205 ± 0.323 | 5.924 ± 0.589 |
7.329 ± 1.735 | 6.426 ± 0.257 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.807 ± 0.327 | 5.823 ± 0.543 |
6.225 ± 0.284 | 4.518 ± 0.739 |
5.321 ± 0.611 | 6.827 ± 0.129 |
6.526 ± 0.893 | 5.522 ± 0.425 |
1.807 ± 0.202 | 3.614 ± 0.253 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
