
Xanthophyllomyces dendrorhous virus L1B
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Totivirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
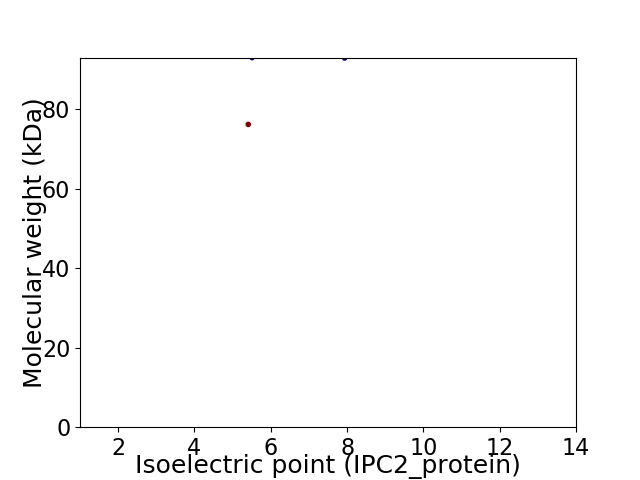
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H9XW62|H9XW62_9VIRU RNA-directed RNA polymerase OS=Xanthophyllomyces dendrorhous virus L1B OX=1167691 GN=RdRp PE=3 SV=1
MM1 pKa = 6.77LTEE4 pKa = 4.15YY5 pKa = 10.86VNSIVRR11 pKa = 11.84LGNGGAFEE19 pKa = 4.23PTLGDD24 pKa = 3.54GRR26 pKa = 11.84FTMVNKK32 pKa = 9.0TRR34 pKa = 11.84AVMEE38 pKa = 4.36LGGVKK43 pKa = 10.02YY44 pKa = 10.8ASNLEE49 pKa = 4.32LATHH53 pKa = 5.9FQVAKK58 pKa = 10.71AKK60 pKa = 10.32FDD62 pKa = 3.8VIKK65 pKa = 10.41PVARR69 pKa = 11.84SNFYY73 pKa = 10.78GYY75 pKa = 10.39NKK77 pKa = 10.25KK78 pKa = 10.58FIDD81 pKa = 3.78EE82 pKa = 4.37SGVYY86 pKa = 10.23DD87 pKa = 4.23PLRR90 pKa = 11.84AIDD93 pKa = 4.02EE94 pKa = 4.38YY95 pKa = 11.52SRR97 pKa = 11.84TVPGMRR103 pKa = 11.84LGRR106 pKa = 11.84DD107 pKa = 3.49DD108 pKa = 5.87LRR110 pKa = 11.84ALSLTDD116 pKa = 3.75NKK118 pKa = 10.32TDD120 pKa = 3.33SQEE123 pKa = 3.87AYY125 pKa = 8.92IYY127 pKa = 11.26NMLVSWLKK135 pKa = 10.33ARR137 pKa = 11.84LYY139 pKa = 11.26VDD141 pKa = 3.92MKK143 pKa = 11.27GSDD146 pKa = 3.09NKK148 pKa = 9.89FTVKK152 pKa = 10.44YY153 pKa = 10.69SSFKK157 pKa = 9.33DD158 pKa = 3.15THH160 pKa = 7.18VGYY163 pKa = 10.34DD164 pKa = 3.01INDD167 pKa = 3.48SYY169 pKa = 11.62GIEE172 pKa = 4.08SVEE175 pKa = 4.38VNLGPPNPTGEE186 pKa = 4.05AAISLEE192 pKa = 4.56FRR194 pKa = 11.84DD195 pKa = 4.76TNNFWSKK202 pKa = 10.73PYY204 pKa = 10.14VLKK207 pKa = 11.02YY208 pKa = 10.63SNNSMEE214 pKa = 3.92QGSFYY219 pKa = 10.39LAHH222 pKa = 6.3VLGSNGTSGLSADD235 pKa = 3.79IQIDD239 pKa = 3.4ALDD242 pKa = 4.01FNEE245 pKa = 5.14LLLDD249 pKa = 3.87PVGSFPTGAFNIFADD264 pKa = 5.79FWAKK268 pKa = 10.63PNVIWLWIMDD278 pKa = 3.86YY279 pKa = 11.19VRR281 pKa = 11.84LNRR284 pKa = 11.84VEE286 pKa = 4.31QEE288 pKa = 3.75FASAFEE294 pKa = 4.32LLGALATQPLPSYY307 pKa = 10.2HH308 pKa = 7.36EE309 pKa = 4.76SILWSKK315 pKa = 10.88SRR317 pKa = 11.84TVVNMSKK324 pKa = 10.62FSPTRR329 pKa = 11.84ARR331 pKa = 11.84VPANLTGEE339 pKa = 4.45PNVHH343 pKa = 7.11DD344 pKa = 5.51LNAQQFTFDD353 pKa = 4.26EE354 pKa = 4.7EE355 pKa = 4.14KK356 pKa = 10.97SPAGFITASAVLNYY370 pKa = 10.25AFWIGIYY377 pKa = 10.93GMVSNFAEE385 pKa = 4.85DD386 pKa = 3.72CSDD389 pKa = 3.04WTDD392 pKa = 3.49AFISSDD398 pKa = 3.62AEE400 pKa = 4.05LGILSTVEE408 pKa = 3.99ARR410 pKa = 11.84PAMISLVTGKK420 pKa = 8.07EE421 pKa = 4.26TNSCFSNNCFLTYY434 pKa = 10.32DD435 pKa = 4.0LSGMYY440 pKa = 9.56GVKK443 pKa = 10.08QLIVDD448 pKa = 3.95EE449 pKa = 5.51KK450 pKa = 10.68IDD452 pKa = 4.06PNHH455 pKa = 6.94PGVILFDD462 pKa = 3.67TVPAFVSGSLLMGAVATDD480 pKa = 3.8YY481 pKa = 10.87PVLKK485 pKa = 10.25HH486 pKa = 6.39LEE488 pKa = 3.99PHH490 pKa = 4.55QHH492 pKa = 5.76IKK494 pKa = 10.32VEE496 pKa = 4.18RR497 pKa = 11.84DD498 pKa = 3.02GLLGARR504 pKa = 11.84EE505 pKa = 4.17AAMLANSYY513 pKa = 10.95RR514 pKa = 11.84LFGNDD519 pKa = 3.15VIIEE523 pKa = 4.16HH524 pKa = 6.09FRR526 pKa = 11.84SAEE529 pKa = 4.06VYY531 pKa = 7.29PTYY534 pKa = 11.08ANSEE538 pKa = 4.22EE539 pKa = 4.6CVIATYY545 pKa = 10.82EE546 pKa = 3.96LFGRR550 pKa = 11.84TRR552 pKa = 11.84TFDD555 pKa = 2.97IMRR558 pKa = 11.84VSSSYY563 pKa = 10.83KK564 pKa = 8.38RR565 pKa = 11.84TGRR568 pKa = 11.84TYY570 pKa = 9.54EE571 pKa = 4.11IPDD574 pKa = 3.44ATHH577 pKa = 4.98VRR579 pKa = 11.84TYY581 pKa = 10.93GEE583 pKa = 4.64CKK585 pKa = 9.48LTMDD589 pKa = 3.96MPVLAVCGWKK599 pKa = 9.38QRR601 pKa = 11.84KK602 pKa = 5.78TVHH605 pKa = 6.1RR606 pKa = 11.84PKK608 pKa = 10.49MMLDD612 pKa = 3.27SRR614 pKa = 11.84RR615 pKa = 11.84VATKK619 pKa = 9.72FMVGATSGFEE629 pKa = 3.76KK630 pKa = 10.47TRR632 pKa = 11.84FAVYY636 pKa = 10.29NRR638 pKa = 11.84RR639 pKa = 11.84NVMAQGFHH647 pKa = 5.4EE648 pKa = 5.39AKK650 pKa = 10.88AEE652 pKa = 3.94MAPALPLVRR661 pKa = 11.84GSAVSTAPIEE671 pKa = 4.09PVQEE675 pKa = 3.99QEE677 pKa = 3.74PANAVEE683 pKa = 4.19
MM1 pKa = 6.77LTEE4 pKa = 4.15YY5 pKa = 10.86VNSIVRR11 pKa = 11.84LGNGGAFEE19 pKa = 4.23PTLGDD24 pKa = 3.54GRR26 pKa = 11.84FTMVNKK32 pKa = 9.0TRR34 pKa = 11.84AVMEE38 pKa = 4.36LGGVKK43 pKa = 10.02YY44 pKa = 10.8ASNLEE49 pKa = 4.32LATHH53 pKa = 5.9FQVAKK58 pKa = 10.71AKK60 pKa = 10.32FDD62 pKa = 3.8VIKK65 pKa = 10.41PVARR69 pKa = 11.84SNFYY73 pKa = 10.78GYY75 pKa = 10.39NKK77 pKa = 10.25KK78 pKa = 10.58FIDD81 pKa = 3.78EE82 pKa = 4.37SGVYY86 pKa = 10.23DD87 pKa = 4.23PLRR90 pKa = 11.84AIDD93 pKa = 4.02EE94 pKa = 4.38YY95 pKa = 11.52SRR97 pKa = 11.84TVPGMRR103 pKa = 11.84LGRR106 pKa = 11.84DD107 pKa = 3.49DD108 pKa = 5.87LRR110 pKa = 11.84ALSLTDD116 pKa = 3.75NKK118 pKa = 10.32TDD120 pKa = 3.33SQEE123 pKa = 3.87AYY125 pKa = 8.92IYY127 pKa = 11.26NMLVSWLKK135 pKa = 10.33ARR137 pKa = 11.84LYY139 pKa = 11.26VDD141 pKa = 3.92MKK143 pKa = 11.27GSDD146 pKa = 3.09NKK148 pKa = 9.89FTVKK152 pKa = 10.44YY153 pKa = 10.69SSFKK157 pKa = 9.33DD158 pKa = 3.15THH160 pKa = 7.18VGYY163 pKa = 10.34DD164 pKa = 3.01INDD167 pKa = 3.48SYY169 pKa = 11.62GIEE172 pKa = 4.08SVEE175 pKa = 4.38VNLGPPNPTGEE186 pKa = 4.05AAISLEE192 pKa = 4.56FRR194 pKa = 11.84DD195 pKa = 4.76TNNFWSKK202 pKa = 10.73PYY204 pKa = 10.14VLKK207 pKa = 11.02YY208 pKa = 10.63SNNSMEE214 pKa = 3.92QGSFYY219 pKa = 10.39LAHH222 pKa = 6.3VLGSNGTSGLSADD235 pKa = 3.79IQIDD239 pKa = 3.4ALDD242 pKa = 4.01FNEE245 pKa = 5.14LLLDD249 pKa = 3.87PVGSFPTGAFNIFADD264 pKa = 5.79FWAKK268 pKa = 10.63PNVIWLWIMDD278 pKa = 3.86YY279 pKa = 11.19VRR281 pKa = 11.84LNRR284 pKa = 11.84VEE286 pKa = 4.31QEE288 pKa = 3.75FASAFEE294 pKa = 4.32LLGALATQPLPSYY307 pKa = 10.2HH308 pKa = 7.36EE309 pKa = 4.76SILWSKK315 pKa = 10.88SRR317 pKa = 11.84TVVNMSKK324 pKa = 10.62FSPTRR329 pKa = 11.84ARR331 pKa = 11.84VPANLTGEE339 pKa = 4.45PNVHH343 pKa = 7.11DD344 pKa = 5.51LNAQQFTFDD353 pKa = 4.26EE354 pKa = 4.7EE355 pKa = 4.14KK356 pKa = 10.97SPAGFITASAVLNYY370 pKa = 10.25AFWIGIYY377 pKa = 10.93GMVSNFAEE385 pKa = 4.85DD386 pKa = 3.72CSDD389 pKa = 3.04WTDD392 pKa = 3.49AFISSDD398 pKa = 3.62AEE400 pKa = 4.05LGILSTVEE408 pKa = 3.99ARR410 pKa = 11.84PAMISLVTGKK420 pKa = 8.07EE421 pKa = 4.26TNSCFSNNCFLTYY434 pKa = 10.32DD435 pKa = 4.0LSGMYY440 pKa = 9.56GVKK443 pKa = 10.08QLIVDD448 pKa = 3.95EE449 pKa = 5.51KK450 pKa = 10.68IDD452 pKa = 4.06PNHH455 pKa = 6.94PGVILFDD462 pKa = 3.67TVPAFVSGSLLMGAVATDD480 pKa = 3.8YY481 pKa = 10.87PVLKK485 pKa = 10.25HH486 pKa = 6.39LEE488 pKa = 3.99PHH490 pKa = 4.55QHH492 pKa = 5.76IKK494 pKa = 10.32VEE496 pKa = 4.18RR497 pKa = 11.84DD498 pKa = 3.02GLLGARR504 pKa = 11.84EE505 pKa = 4.17AAMLANSYY513 pKa = 10.95RR514 pKa = 11.84LFGNDD519 pKa = 3.15VIIEE523 pKa = 4.16HH524 pKa = 6.09FRR526 pKa = 11.84SAEE529 pKa = 4.06VYY531 pKa = 7.29PTYY534 pKa = 11.08ANSEE538 pKa = 4.22EE539 pKa = 4.6CVIATYY545 pKa = 10.82EE546 pKa = 3.96LFGRR550 pKa = 11.84TRR552 pKa = 11.84TFDD555 pKa = 2.97IMRR558 pKa = 11.84VSSSYY563 pKa = 10.83KK564 pKa = 8.38RR565 pKa = 11.84TGRR568 pKa = 11.84TYY570 pKa = 9.54EE571 pKa = 4.11IPDD574 pKa = 3.44ATHH577 pKa = 4.98VRR579 pKa = 11.84TYY581 pKa = 10.93GEE583 pKa = 4.64CKK585 pKa = 9.48LTMDD589 pKa = 3.96MPVLAVCGWKK599 pKa = 9.38QRR601 pKa = 11.84KK602 pKa = 5.78TVHH605 pKa = 6.1RR606 pKa = 11.84PKK608 pKa = 10.49MMLDD612 pKa = 3.27SRR614 pKa = 11.84RR615 pKa = 11.84VATKK619 pKa = 9.72FMVGATSGFEE629 pKa = 3.76KK630 pKa = 10.47TRR632 pKa = 11.84FAVYY636 pKa = 10.29NRR638 pKa = 11.84RR639 pKa = 11.84NVMAQGFHH647 pKa = 5.4EE648 pKa = 5.39AKK650 pKa = 10.88AEE652 pKa = 3.94MAPALPLVRR661 pKa = 11.84GSAVSTAPIEE671 pKa = 4.09PVQEE675 pKa = 3.99QEE677 pKa = 3.74PANAVEE683 pKa = 4.19
Molecular weight: 76.22 kDa
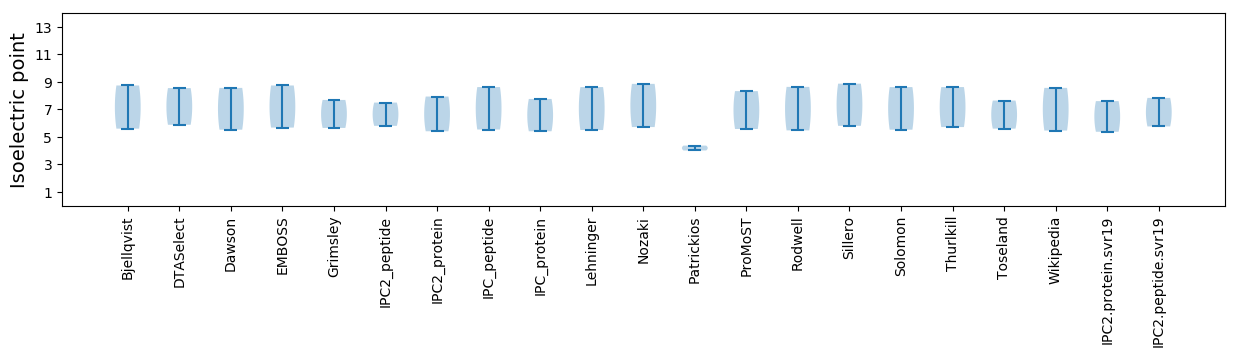
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9XW62|H9XW62_9VIRU RNA-directed RNA polymerase OS=Xanthophyllomyces dendrorhous virus L1B OX=1167691 GN=RdRp PE=3 SV=1
MM1 pKa = 7.15TSEE4 pKa = 4.63SVQHH8 pKa = 6.44CEE10 pKa = 3.7EE11 pKa = 4.32LNGIKK16 pKa = 9.79IIKK19 pKa = 9.07SVRR22 pKa = 11.84NKK24 pKa = 9.87AMKK27 pKa = 9.44MEE29 pKa = 4.04MRR31 pKa = 11.84DD32 pKa = 3.77GYY34 pKa = 11.83VPMCIKK40 pKa = 10.37ARR42 pKa = 11.84PGIVYY47 pKa = 9.41KK48 pKa = 10.5WEE50 pKa = 3.78ATAFCDD56 pKa = 4.28SEE58 pKa = 4.57YY59 pKa = 11.42VLVGSDD65 pKa = 3.06MSEE68 pKa = 3.81PSEE71 pKa = 4.11NEE73 pKa = 3.84SNVNIMGVLMSGLRR87 pKa = 11.84LAGDD91 pKa = 3.48NCTYY95 pKa = 10.97LYY97 pKa = 11.23AKK99 pKa = 9.77VDD101 pKa = 3.47QYY103 pKa = 12.05LVTRR107 pKa = 11.84KK108 pKa = 9.68KK109 pKa = 10.69AVIAIMTRR117 pKa = 11.84HH118 pKa = 5.5FNGLYY123 pKa = 10.67GSVFLNDD130 pKa = 4.41PLNMEE135 pKa = 4.06AMFRR139 pKa = 11.84DD140 pKa = 3.87RR141 pKa = 11.84APDD144 pKa = 3.74DD145 pKa = 3.65EE146 pKa = 5.97LMPKK150 pKa = 9.88QNYY153 pKa = 8.84EE154 pKa = 4.25KK155 pKa = 10.18ICKK158 pKa = 9.74KK159 pKa = 10.55EE160 pKa = 3.81NTKK163 pKa = 8.48ITAQHH168 pKa = 6.64HH169 pKa = 4.49IHH171 pKa = 5.78FTSEE175 pKa = 3.62EE176 pKa = 4.19VVRR179 pKa = 11.84VLGKK183 pKa = 8.95RR184 pKa = 11.84TAEE187 pKa = 3.77EE188 pKa = 4.3SEE190 pKa = 4.27ATRR193 pKa = 11.84LPADD197 pKa = 3.38ATMSMAAGVLLWYY210 pKa = 10.76NEE212 pKa = 3.91LSPVLKK218 pKa = 10.66EE219 pKa = 4.39MILKK223 pKa = 10.65CGLFKK228 pKa = 11.12SKK230 pKa = 9.6TIQAFKK236 pKa = 10.65KK237 pKa = 8.1IAKK240 pKa = 9.47DD241 pKa = 3.08ISVEE245 pKa = 4.06AKK247 pKa = 9.59SLQNIVQTDD256 pKa = 3.58LRR258 pKa = 11.84SVFEE262 pKa = 4.61IDD264 pKa = 3.28TLINRR269 pKa = 11.84IDD271 pKa = 4.25GEE273 pKa = 4.32VDD275 pKa = 2.53WAEE278 pKa = 4.16EE279 pKa = 3.62RR280 pKa = 11.84DD281 pKa = 3.72HH282 pKa = 6.98RR283 pKa = 11.84VNPNVTNLSYY293 pKa = 11.44SDD295 pKa = 4.08VYY297 pKa = 11.25DD298 pKa = 3.66AAKK301 pKa = 10.61DD302 pKa = 3.45IFLQAAAVGRR312 pKa = 11.84KK313 pKa = 7.88PVSMDD318 pKa = 2.47WDD320 pKa = 4.17KK321 pKa = 11.57YY322 pKa = 8.65WASRR326 pKa = 11.84WQWSAAGSIHH336 pKa = 5.44SQYY339 pKa = 11.04VEE341 pKa = 3.83DD342 pKa = 4.27DD343 pKa = 3.75KK344 pKa = 11.97YY345 pKa = 11.59VIRR348 pKa = 11.84TDD350 pKa = 3.68RR351 pKa = 11.84NLKK354 pKa = 10.3NKK356 pKa = 10.12FIAIANMPKK365 pKa = 9.93YY366 pKa = 10.56GYY368 pKa = 10.58DD369 pKa = 3.04FFMSRR374 pKa = 11.84KK375 pKa = 7.8PQMHH379 pKa = 6.53AWASIKK385 pKa = 10.37YY386 pKa = 8.37EE387 pKa = 3.89WGKK390 pKa = 10.34LRR392 pKa = 11.84AIYY395 pKa = 8.57GTDD398 pKa = 2.92LTSYY402 pKa = 10.13VLSNFAFYY410 pKa = 10.89NCEE413 pKa = 3.54NVLPKK418 pKa = 10.28RR419 pKa = 11.84FPVGKK424 pKa = 9.66DD425 pKa = 2.98ANDD428 pKa = 3.55ANVVNRR434 pKa = 11.84VAGVLKK440 pKa = 10.75DD441 pKa = 3.78RR442 pKa = 11.84LPYY445 pKa = 10.8CLDD448 pKa = 3.63FEE450 pKa = 5.44DD451 pKa = 5.6FNSQHH456 pKa = 6.36SVSSMKK462 pKa = 10.66AVIYY466 pKa = 10.14AYY468 pKa = 10.16GDD470 pKa = 3.32VYY472 pKa = 10.91RR473 pKa = 11.84QVFTEE478 pKa = 3.96QQLEE482 pKa = 4.1ALAWTAEE489 pKa = 4.11SLDD492 pKa = 3.7DD493 pKa = 4.67VSVNDD498 pKa = 4.18NVGLKK503 pKa = 7.8QTYY506 pKa = 9.64KK507 pKa = 11.12SNATLLSGWRR517 pKa = 11.84LTTFVNSVLNAVYY530 pKa = 9.7TDD532 pKa = 4.69KK533 pKa = 11.13ICGEE537 pKa = 4.02AKK539 pKa = 9.81MPGSSLHH546 pKa = 6.44NGDD549 pKa = 4.77DD550 pKa = 3.75VLIGATSMKK559 pKa = 9.81VARR562 pKa = 11.84EE563 pKa = 3.75SLRR566 pKa = 11.84RR567 pKa = 11.84SEE569 pKa = 4.65LYY571 pKa = 10.55GIRR574 pKa = 11.84VQASKK579 pKa = 10.92CAFGGIAEE587 pKa = 4.67FLRR590 pKa = 11.84IDD592 pKa = 3.45HH593 pKa = 6.7ARR595 pKa = 11.84GSKK598 pKa = 9.72GQYY601 pKa = 6.9LTRR604 pKa = 11.84AIATLMHH611 pKa = 6.29SRR613 pKa = 11.84IEE615 pKa = 4.5SKK617 pKa = 10.86LSTDD621 pKa = 3.22ARR623 pKa = 11.84DD624 pKa = 3.81LIEE627 pKa = 4.82AMEE630 pKa = 4.28NRR632 pKa = 11.84FSDD635 pKa = 3.83CLNRR639 pKa = 11.84GMTLDD644 pKa = 3.37TVTKK648 pKa = 10.21LRR650 pKa = 11.84HH651 pKa = 4.95VYY653 pKa = 10.26YY654 pKa = 10.45NRR656 pKa = 11.84QSVICNMPVEE666 pKa = 4.28DD667 pKa = 5.2FYY669 pKa = 11.35RR670 pKa = 11.84IKK672 pKa = 8.18TTHH675 pKa = 6.14RR676 pKa = 11.84VAGGVSEE683 pKa = 6.02AIDD686 pKa = 3.81SDD688 pKa = 3.67VSMSVIRR695 pKa = 11.84GIDD698 pKa = 2.94KK699 pKa = 10.84SFNIKK704 pKa = 9.61IPKK707 pKa = 8.69LQGVHH712 pKa = 7.11DD713 pKa = 4.27YY714 pKa = 11.17ARR716 pKa = 11.84AVAMEE721 pKa = 4.41LEE723 pKa = 4.22MMKK726 pKa = 10.53RR727 pKa = 11.84LDD729 pKa = 4.09YY730 pKa = 10.52ITDD733 pKa = 3.32RR734 pKa = 11.84MYY736 pKa = 10.98KK737 pKa = 8.07ATYY740 pKa = 8.66EE741 pKa = 4.03AVVPKK746 pKa = 9.62TRR748 pKa = 11.84GMTVVPNKK756 pKa = 9.42EE757 pKa = 4.16EE758 pKa = 3.77KK759 pKa = 8.73WCYY762 pKa = 8.5NVKK765 pKa = 10.48AIYY768 pKa = 9.86KK769 pKa = 9.08AFKK772 pKa = 10.75GKK774 pKa = 9.57IQTAGYY780 pKa = 9.69GKK782 pKa = 10.41AALVGMALDD791 pKa = 4.92VIQTSDD797 pKa = 3.29KK798 pKa = 10.87DD799 pKa = 3.86TTLKK803 pKa = 9.92MALARR808 pKa = 11.84SPDD811 pKa = 4.24PIKK814 pKa = 10.66LLRR817 pKa = 11.84YY818 pKa = 9.24LVV820 pKa = 3.53
MM1 pKa = 7.15TSEE4 pKa = 4.63SVQHH8 pKa = 6.44CEE10 pKa = 3.7EE11 pKa = 4.32LNGIKK16 pKa = 9.79IIKK19 pKa = 9.07SVRR22 pKa = 11.84NKK24 pKa = 9.87AMKK27 pKa = 9.44MEE29 pKa = 4.04MRR31 pKa = 11.84DD32 pKa = 3.77GYY34 pKa = 11.83VPMCIKK40 pKa = 10.37ARR42 pKa = 11.84PGIVYY47 pKa = 9.41KK48 pKa = 10.5WEE50 pKa = 3.78ATAFCDD56 pKa = 4.28SEE58 pKa = 4.57YY59 pKa = 11.42VLVGSDD65 pKa = 3.06MSEE68 pKa = 3.81PSEE71 pKa = 4.11NEE73 pKa = 3.84SNVNIMGVLMSGLRR87 pKa = 11.84LAGDD91 pKa = 3.48NCTYY95 pKa = 10.97LYY97 pKa = 11.23AKK99 pKa = 9.77VDD101 pKa = 3.47QYY103 pKa = 12.05LVTRR107 pKa = 11.84KK108 pKa = 9.68KK109 pKa = 10.69AVIAIMTRR117 pKa = 11.84HH118 pKa = 5.5FNGLYY123 pKa = 10.67GSVFLNDD130 pKa = 4.41PLNMEE135 pKa = 4.06AMFRR139 pKa = 11.84DD140 pKa = 3.87RR141 pKa = 11.84APDD144 pKa = 3.74DD145 pKa = 3.65EE146 pKa = 5.97LMPKK150 pKa = 9.88QNYY153 pKa = 8.84EE154 pKa = 4.25KK155 pKa = 10.18ICKK158 pKa = 9.74KK159 pKa = 10.55EE160 pKa = 3.81NTKK163 pKa = 8.48ITAQHH168 pKa = 6.64HH169 pKa = 4.49IHH171 pKa = 5.78FTSEE175 pKa = 3.62EE176 pKa = 4.19VVRR179 pKa = 11.84VLGKK183 pKa = 8.95RR184 pKa = 11.84TAEE187 pKa = 3.77EE188 pKa = 4.3SEE190 pKa = 4.27ATRR193 pKa = 11.84LPADD197 pKa = 3.38ATMSMAAGVLLWYY210 pKa = 10.76NEE212 pKa = 3.91LSPVLKK218 pKa = 10.66EE219 pKa = 4.39MILKK223 pKa = 10.65CGLFKK228 pKa = 11.12SKK230 pKa = 9.6TIQAFKK236 pKa = 10.65KK237 pKa = 8.1IAKK240 pKa = 9.47DD241 pKa = 3.08ISVEE245 pKa = 4.06AKK247 pKa = 9.59SLQNIVQTDD256 pKa = 3.58LRR258 pKa = 11.84SVFEE262 pKa = 4.61IDD264 pKa = 3.28TLINRR269 pKa = 11.84IDD271 pKa = 4.25GEE273 pKa = 4.32VDD275 pKa = 2.53WAEE278 pKa = 4.16EE279 pKa = 3.62RR280 pKa = 11.84DD281 pKa = 3.72HH282 pKa = 6.98RR283 pKa = 11.84VNPNVTNLSYY293 pKa = 11.44SDD295 pKa = 4.08VYY297 pKa = 11.25DD298 pKa = 3.66AAKK301 pKa = 10.61DD302 pKa = 3.45IFLQAAAVGRR312 pKa = 11.84KK313 pKa = 7.88PVSMDD318 pKa = 2.47WDD320 pKa = 4.17KK321 pKa = 11.57YY322 pKa = 8.65WASRR326 pKa = 11.84WQWSAAGSIHH336 pKa = 5.44SQYY339 pKa = 11.04VEE341 pKa = 3.83DD342 pKa = 4.27DD343 pKa = 3.75KK344 pKa = 11.97YY345 pKa = 11.59VIRR348 pKa = 11.84TDD350 pKa = 3.68RR351 pKa = 11.84NLKK354 pKa = 10.3NKK356 pKa = 10.12FIAIANMPKK365 pKa = 9.93YY366 pKa = 10.56GYY368 pKa = 10.58DD369 pKa = 3.04FFMSRR374 pKa = 11.84KK375 pKa = 7.8PQMHH379 pKa = 6.53AWASIKK385 pKa = 10.37YY386 pKa = 8.37EE387 pKa = 3.89WGKK390 pKa = 10.34LRR392 pKa = 11.84AIYY395 pKa = 8.57GTDD398 pKa = 2.92LTSYY402 pKa = 10.13VLSNFAFYY410 pKa = 10.89NCEE413 pKa = 3.54NVLPKK418 pKa = 10.28RR419 pKa = 11.84FPVGKK424 pKa = 9.66DD425 pKa = 2.98ANDD428 pKa = 3.55ANVVNRR434 pKa = 11.84VAGVLKK440 pKa = 10.75DD441 pKa = 3.78RR442 pKa = 11.84LPYY445 pKa = 10.8CLDD448 pKa = 3.63FEE450 pKa = 5.44DD451 pKa = 5.6FNSQHH456 pKa = 6.36SVSSMKK462 pKa = 10.66AVIYY466 pKa = 10.14AYY468 pKa = 10.16GDD470 pKa = 3.32VYY472 pKa = 10.91RR473 pKa = 11.84QVFTEE478 pKa = 3.96QQLEE482 pKa = 4.1ALAWTAEE489 pKa = 4.11SLDD492 pKa = 3.7DD493 pKa = 4.67VSVNDD498 pKa = 4.18NVGLKK503 pKa = 7.8QTYY506 pKa = 9.64KK507 pKa = 11.12SNATLLSGWRR517 pKa = 11.84LTTFVNSVLNAVYY530 pKa = 9.7TDD532 pKa = 4.69KK533 pKa = 11.13ICGEE537 pKa = 4.02AKK539 pKa = 9.81MPGSSLHH546 pKa = 6.44NGDD549 pKa = 4.77DD550 pKa = 3.75VLIGATSMKK559 pKa = 9.81VARR562 pKa = 11.84EE563 pKa = 3.75SLRR566 pKa = 11.84RR567 pKa = 11.84SEE569 pKa = 4.65LYY571 pKa = 10.55GIRR574 pKa = 11.84VQASKK579 pKa = 10.92CAFGGIAEE587 pKa = 4.67FLRR590 pKa = 11.84IDD592 pKa = 3.45HH593 pKa = 6.7ARR595 pKa = 11.84GSKK598 pKa = 9.72GQYY601 pKa = 6.9LTRR604 pKa = 11.84AIATLMHH611 pKa = 6.29SRR613 pKa = 11.84IEE615 pKa = 4.5SKK617 pKa = 10.86LSTDD621 pKa = 3.22ARR623 pKa = 11.84DD624 pKa = 3.81LIEE627 pKa = 4.82AMEE630 pKa = 4.28NRR632 pKa = 11.84FSDD635 pKa = 3.83CLNRR639 pKa = 11.84GMTLDD644 pKa = 3.37TVTKK648 pKa = 10.21LRR650 pKa = 11.84HH651 pKa = 4.95VYY653 pKa = 10.26YY654 pKa = 10.45NRR656 pKa = 11.84QSVICNMPVEE666 pKa = 4.28DD667 pKa = 5.2FYY669 pKa = 11.35RR670 pKa = 11.84IKK672 pKa = 8.18TTHH675 pKa = 6.14RR676 pKa = 11.84VAGGVSEE683 pKa = 6.02AIDD686 pKa = 3.81SDD688 pKa = 3.67VSMSVIRR695 pKa = 11.84GIDD698 pKa = 2.94KK699 pKa = 10.84SFNIKK704 pKa = 9.61IPKK707 pKa = 8.69LQGVHH712 pKa = 7.11DD713 pKa = 4.27YY714 pKa = 11.17ARR716 pKa = 11.84AVAMEE721 pKa = 4.41LEE723 pKa = 4.22MMKK726 pKa = 10.53RR727 pKa = 11.84LDD729 pKa = 4.09YY730 pKa = 10.52ITDD733 pKa = 3.32RR734 pKa = 11.84MYY736 pKa = 10.98KK737 pKa = 8.07ATYY740 pKa = 8.66EE741 pKa = 4.03AVVPKK746 pKa = 9.62TRR748 pKa = 11.84GMTVVPNKK756 pKa = 9.42EE757 pKa = 4.16EE758 pKa = 3.77KK759 pKa = 8.73WCYY762 pKa = 8.5NVKK765 pKa = 10.48AIYY768 pKa = 9.86KK769 pKa = 9.08AFKK772 pKa = 10.75GKK774 pKa = 9.57IQTAGYY780 pKa = 9.69GKK782 pKa = 10.41AALVGMALDD791 pKa = 4.92VIQTSDD797 pKa = 3.29KK798 pKa = 10.87DD799 pKa = 3.86TTLKK803 pKa = 9.92MALARR808 pKa = 11.84SPDD811 pKa = 4.24PIKK814 pKa = 10.66LLRR817 pKa = 11.84YY818 pKa = 9.24LVV820 pKa = 3.53
Molecular weight: 92.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1503 |
683 |
820 |
751.5 |
84.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.649 ± 0.007 | 1.264 ± 0.247 |
6.121 ± 0.357 | 5.855 ± 0.189 |
4.125 ± 0.828 | 5.788 ± 0.512 |
1.863 ± 0.026 | 5.123 ± 0.468 |
6.321 ± 1.047 | 7.784 ± 0.172 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.726 ± 0.323 | 5.123 ± 0.188 |
3.593 ± 0.7 | 2.395 ± 0.221 |
5.522 ± 0.255 | 7.252 ± 0.231 |
5.522 ± 0.308 | 7.984 ± 0.05 |
1.397 ± 0.051 | 4.591 ± 0.127 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
