
Prevotella baroniae F0067
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Prevotellaceae; Prevotella; Prevotella baroniae
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
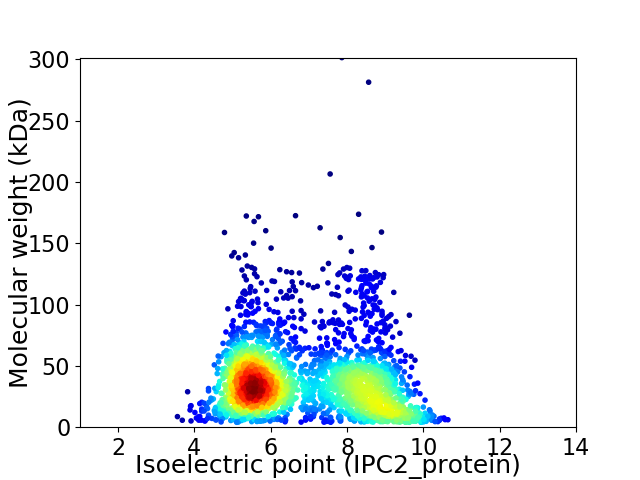
Virtual 2D-PAGE plot for 2480 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U2NLW6|U2NLW6_9BACT Putative lipoprotein OS=Prevotella baroniae F0067 OX=1115809 GN=HMPREF9135_1221 PE=4 SV=1
MM1 pKa = 7.33SVFISKK7 pKa = 10.09EE8 pKa = 4.07LIPKK12 pKa = 9.56IEE14 pKa = 3.97RR15 pKa = 11.84YY16 pKa = 10.32YY17 pKa = 9.87NIKK20 pKa = 8.13MTDD23 pKa = 3.18PTEE26 pKa = 3.81EE27 pKa = 3.72VGYY30 pKa = 10.64GEE32 pKa = 3.84QAMAFEE38 pKa = 5.7DD39 pKa = 4.12VTDD42 pKa = 3.86SCSDD46 pKa = 3.61LEE48 pKa = 4.3DD49 pKa = 3.27WLEE52 pKa = 4.26NGDD55 pKa = 4.43VLTCYY60 pKa = 10.49EE61 pKa = 4.44DD62 pKa = 4.24ADD64 pKa = 4.04GQIHH68 pKa = 5.93YY69 pKa = 9.82KK70 pKa = 10.4VSVEE74 pKa = 3.82NGVFDD79 pKa = 4.82FVLDD83 pKa = 3.9KK84 pKa = 11.54EE85 pKa = 4.39NVEE88 pKa = 4.17YY89 pKa = 10.57EE90 pKa = 4.48SYY92 pKa = 11.36LEE94 pKa = 5.39DD95 pKa = 4.72IKK97 pKa = 11.63SDD99 pKa = 3.8LNNLFSNASVEE110 pKa = 4.56DD111 pKa = 4.35FRR113 pKa = 11.84TLCQNLQDD121 pKa = 3.56YY122 pKa = 11.29DD123 pKa = 3.97EE124 pKa = 4.62YY125 pKa = 11.29KK126 pKa = 10.47KK127 pKa = 10.7SHH129 pKa = 6.77EE130 pKa = 4.16DD131 pKa = 3.48VIDD134 pKa = 3.91FDD136 pKa = 6.24LIDD139 pKa = 4.16KK140 pKa = 10.31EE141 pKa = 4.52DD142 pKa = 3.66YY143 pKa = 10.76EE144 pKa = 5.51SIVKK148 pKa = 9.62EE149 pKa = 4.37AITEE153 pKa = 4.18YY154 pKa = 10.46IEE156 pKa = 4.73KK157 pKa = 10.47NDD159 pKa = 3.11IKK161 pKa = 11.36AEE163 pKa = 4.01IKK165 pKa = 10.61GLSADD170 pKa = 4.04DD171 pKa = 3.76VVMDD175 pKa = 4.5GDD177 pKa = 4.02NSFTYY182 pKa = 10.04KK183 pKa = 10.72GEE185 pKa = 4.01EE186 pKa = 3.94YY187 pKa = 10.54QGFDD191 pKa = 3.71SSDD194 pKa = 3.27GGDD197 pKa = 4.01FDD199 pKa = 4.93CTSCEE204 pKa = 4.01NFDD207 pKa = 5.01LIYY210 pKa = 10.74EE211 pKa = 4.78AVQEE215 pKa = 4.37ANCEE219 pKa = 4.17DD220 pKa = 4.11KK221 pKa = 11.5EE222 pKa = 4.21EE223 pKa = 4.02LTMYY227 pKa = 10.16LCGMNFVYY235 pKa = 10.56KK236 pKa = 10.78NLVDD240 pKa = 3.55DD241 pKa = 4.27VMYY244 pKa = 10.94KK245 pKa = 10.29FYY247 pKa = 11.04FKK249 pKa = 11.09
MM1 pKa = 7.33SVFISKK7 pKa = 10.09EE8 pKa = 4.07LIPKK12 pKa = 9.56IEE14 pKa = 3.97RR15 pKa = 11.84YY16 pKa = 10.32YY17 pKa = 9.87NIKK20 pKa = 8.13MTDD23 pKa = 3.18PTEE26 pKa = 3.81EE27 pKa = 3.72VGYY30 pKa = 10.64GEE32 pKa = 3.84QAMAFEE38 pKa = 5.7DD39 pKa = 4.12VTDD42 pKa = 3.86SCSDD46 pKa = 3.61LEE48 pKa = 4.3DD49 pKa = 3.27WLEE52 pKa = 4.26NGDD55 pKa = 4.43VLTCYY60 pKa = 10.49EE61 pKa = 4.44DD62 pKa = 4.24ADD64 pKa = 4.04GQIHH68 pKa = 5.93YY69 pKa = 9.82KK70 pKa = 10.4VSVEE74 pKa = 3.82NGVFDD79 pKa = 4.82FVLDD83 pKa = 3.9KK84 pKa = 11.54EE85 pKa = 4.39NVEE88 pKa = 4.17YY89 pKa = 10.57EE90 pKa = 4.48SYY92 pKa = 11.36LEE94 pKa = 5.39DD95 pKa = 4.72IKK97 pKa = 11.63SDD99 pKa = 3.8LNNLFSNASVEE110 pKa = 4.56DD111 pKa = 4.35FRR113 pKa = 11.84TLCQNLQDD121 pKa = 3.56YY122 pKa = 11.29DD123 pKa = 3.97EE124 pKa = 4.62YY125 pKa = 11.29KK126 pKa = 10.47KK127 pKa = 10.7SHH129 pKa = 6.77EE130 pKa = 4.16DD131 pKa = 3.48VIDD134 pKa = 3.91FDD136 pKa = 6.24LIDD139 pKa = 4.16KK140 pKa = 10.31EE141 pKa = 4.52DD142 pKa = 3.66YY143 pKa = 10.76EE144 pKa = 5.51SIVKK148 pKa = 9.62EE149 pKa = 4.37AITEE153 pKa = 4.18YY154 pKa = 10.46IEE156 pKa = 4.73KK157 pKa = 10.47NDD159 pKa = 3.11IKK161 pKa = 11.36AEE163 pKa = 4.01IKK165 pKa = 10.61GLSADD170 pKa = 4.04DD171 pKa = 3.76VVMDD175 pKa = 4.5GDD177 pKa = 4.02NSFTYY182 pKa = 10.04KK183 pKa = 10.72GEE185 pKa = 4.01EE186 pKa = 3.94YY187 pKa = 10.54QGFDD191 pKa = 3.71SSDD194 pKa = 3.27GGDD197 pKa = 4.01FDD199 pKa = 4.93CTSCEE204 pKa = 4.01NFDD207 pKa = 5.01LIYY210 pKa = 10.74EE211 pKa = 4.78AVQEE215 pKa = 4.37ANCEE219 pKa = 4.17DD220 pKa = 4.11KK221 pKa = 11.5EE222 pKa = 4.21EE223 pKa = 4.02LTMYY227 pKa = 10.16LCGMNFVYY235 pKa = 10.56KK236 pKa = 10.78NLVDD240 pKa = 3.55DD241 pKa = 4.27VMYY244 pKa = 10.94KK245 pKa = 10.29FYY247 pKa = 11.04FKK249 pKa = 11.09
Molecular weight: 28.9 kDa
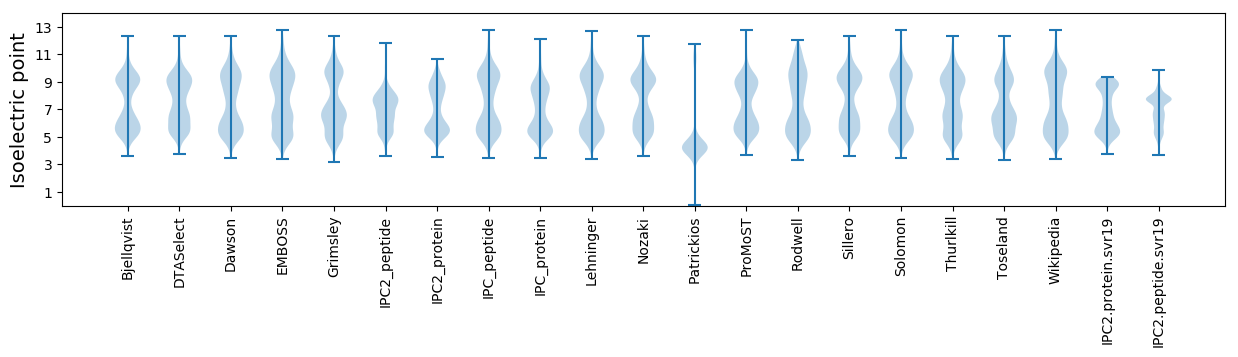
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U2NLX0|U2NLX0_9BACT Acyltransferase OS=Prevotella baroniae F0067 OX=1115809 GN=HMPREF9135_1243 PE=4 SV=1
MM1 pKa = 6.72EE2 pKa = 4.32TVVTLMMLLTVGCFLLKK19 pKa = 8.79QTFMGVRR26 pKa = 11.84QILVTAVVVMMFVALMWPFAITQSKK51 pKa = 6.73TQIASWLSNPQLMLDD66 pKa = 3.77VAVLLSVDD74 pKa = 3.23IALEE78 pKa = 3.72IAFCVTDD85 pKa = 3.19VDD87 pKa = 3.85VRR89 pKa = 11.84TSRR92 pKa = 11.84KK93 pKa = 7.3VSRR96 pKa = 11.84RR97 pKa = 11.84KK98 pKa = 10.29RR99 pKa = 11.84MWFRR103 pKa = 11.84FLRR106 pKa = 11.84HH107 pKa = 5.71FPGLLVFPVFFAVLVWAIFALPGVDD132 pKa = 3.96FALVGWSLGVALLFVLPLLAYY153 pKa = 10.2LLRR156 pKa = 11.84RR157 pKa = 11.84LVPEE161 pKa = 3.8EE162 pKa = 3.86EE163 pKa = 4.39LRR165 pKa = 11.84LEE167 pKa = 5.39LLFLCNLLLGGLGVIATVNGTTAVKK192 pKa = 10.39GVSQVNYY199 pKa = 9.93AALGAMAALILVVGGVGFVARR220 pKa = 11.84RR221 pKa = 11.84VRR223 pKa = 11.84QKK225 pKa = 10.57RR226 pKa = 11.84AVARR230 pKa = 11.84LQRR233 pKa = 11.84KK234 pKa = 8.3QYY236 pKa = 10.76GRR238 pKa = 11.84TACPSHH244 pKa = 6.95PP245 pKa = 3.81
MM1 pKa = 6.72EE2 pKa = 4.32TVVTLMMLLTVGCFLLKK19 pKa = 8.79QTFMGVRR26 pKa = 11.84QILVTAVVVMMFVALMWPFAITQSKK51 pKa = 6.73TQIASWLSNPQLMLDD66 pKa = 3.77VAVLLSVDD74 pKa = 3.23IALEE78 pKa = 3.72IAFCVTDD85 pKa = 3.19VDD87 pKa = 3.85VRR89 pKa = 11.84TSRR92 pKa = 11.84KK93 pKa = 7.3VSRR96 pKa = 11.84RR97 pKa = 11.84KK98 pKa = 10.29RR99 pKa = 11.84MWFRR103 pKa = 11.84FLRR106 pKa = 11.84HH107 pKa = 5.71FPGLLVFPVFFAVLVWAIFALPGVDD132 pKa = 3.96FALVGWSLGVALLFVLPLLAYY153 pKa = 10.2LLRR156 pKa = 11.84RR157 pKa = 11.84LVPEE161 pKa = 3.8EE162 pKa = 3.86EE163 pKa = 4.39LRR165 pKa = 11.84LEE167 pKa = 5.39LLFLCNLLLGGLGVIATVNGTTAVKK192 pKa = 10.39GVSQVNYY199 pKa = 9.93AALGAMAALILVVGGVGFVARR220 pKa = 11.84RR221 pKa = 11.84VRR223 pKa = 11.84QKK225 pKa = 10.57RR226 pKa = 11.84AVARR230 pKa = 11.84LQRR233 pKa = 11.84KK234 pKa = 8.3QYY236 pKa = 10.76GRR238 pKa = 11.84TACPSHH244 pKa = 6.95PP245 pKa = 3.81
Molecular weight: 27.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
896784 |
37 |
2721 |
361.6 |
40.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.934 ± 0.045 | 1.217 ± 0.018 |
5.877 ± 0.034 | 5.79 ± 0.044 |
4.434 ± 0.028 | 7.189 ± 0.043 |
2.154 ± 0.023 | 5.736 ± 0.042 |
6.129 ± 0.041 | 9.12 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.88 ± 0.022 | 4.773 ± 0.041 |
3.763 ± 0.025 | 3.631 ± 0.027 |
5.504 ± 0.04 | 5.838 ± 0.04 |
5.722 ± 0.036 | 6.787 ± 0.038 |
1.281 ± 0.018 | 4.242 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
