
Lake Sarah-associated circular virus-33
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.92
Get precalculated fractions of proteins
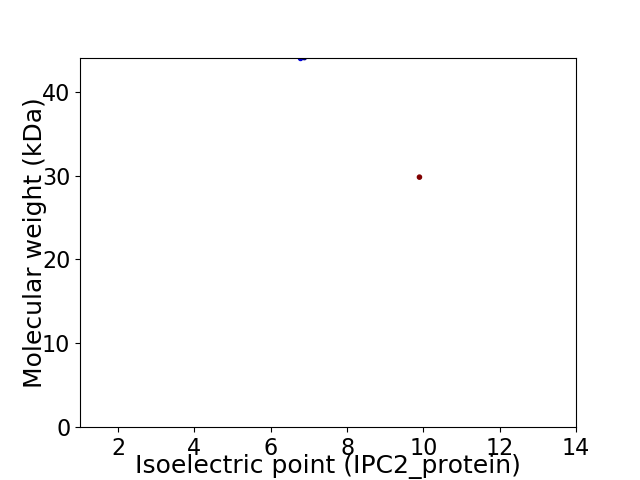
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
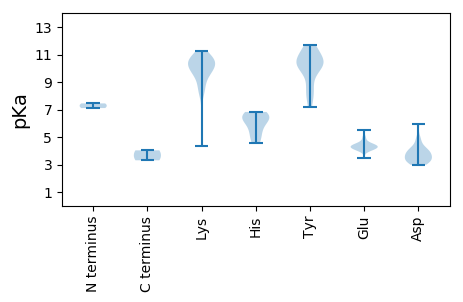
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GAB3|A0A126GAB3_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-33 OX=1685761 PE=4 SV=1
MM1 pKa = 7.45NNNEE5 pKa = 4.12EE6 pKa = 4.21YY7 pKa = 10.59EE8 pKa = 4.01RR9 pKa = 11.84WLNMDD14 pKa = 3.78TFYY17 pKa = 11.67NPLEE21 pKa = 4.04YY22 pKa = 10.51SQTFTPPLSLTTSQQLPPIPMPMTPIEE49 pKa = 4.31GDD51 pKa = 2.99PRR53 pKa = 11.84EE54 pKa = 4.39RR55 pKa = 11.84VGTTAMCGLPRR66 pKa = 11.84AMCAGAPEE74 pKa = 4.17EE75 pKa = 4.46EE76 pKa = 4.42KK77 pKa = 10.94LASAVCALSLEE88 pKa = 4.35DD89 pKa = 4.6LADD92 pKa = 3.61NKK94 pKa = 9.78EE95 pKa = 4.35TFKK98 pKa = 10.97CRR100 pKa = 11.84SIVFTCYY107 pKa = 9.95KK108 pKa = 10.17RR109 pKa = 11.84EE110 pKa = 4.4AIDD113 pKa = 5.93DD114 pKa = 3.93PEE116 pKa = 5.0AWLRR120 pKa = 11.84GLQSKK125 pKa = 8.03CVWVLGQLEE134 pKa = 4.74KK135 pKa = 10.98CPKK138 pKa = 6.67TQRR141 pKa = 11.84IHH143 pKa = 5.39IQGMAYY149 pKa = 10.11NKK151 pKa = 10.42SDD153 pKa = 3.83IAWAMLKK160 pKa = 10.79GEE162 pKa = 4.5STWKK166 pKa = 9.78RR167 pKa = 11.84KK168 pKa = 9.76CIAPIKK174 pKa = 9.85SIEE177 pKa = 4.22YY178 pKa = 7.21CTKK181 pKa = 10.74AKK183 pKa = 10.46SQLAGPWEE191 pKa = 4.62FGDD194 pKa = 3.81RR195 pKa = 11.84PAYY198 pKa = 9.3GQSAKK203 pKa = 9.34TSKK206 pKa = 10.82LNEE209 pKa = 4.27KK210 pKa = 10.71KK211 pKa = 10.69EE212 pKa = 4.11MNATLLTMTPYY223 pKa = 10.35EE224 pKa = 4.53AVDD227 pKa = 3.12EE228 pKa = 5.52GYY230 pKa = 10.37ISWRR234 pKa = 11.84DD235 pKa = 3.54YY236 pKa = 11.54EE237 pKa = 4.24KK238 pKa = 11.25LKK240 pKa = 11.0LFQTTYY246 pKa = 11.07KK247 pKa = 10.19ADD249 pKa = 3.42KK250 pKa = 10.45KK251 pKa = 10.38IAEE254 pKa = 4.19HH255 pKa = 6.47LKK257 pKa = 10.58RR258 pKa = 11.84PIDD261 pKa = 3.26HH262 pKa = 6.82EE263 pKa = 4.39MRR265 pKa = 11.84HH266 pKa = 5.7EE267 pKa = 4.98WIQGKK272 pKa = 8.8SGKK275 pKa = 10.59GKK277 pKa = 4.37TTKK280 pKa = 10.62AILEE284 pKa = 4.47NPDD287 pKa = 4.19HH288 pKa = 6.24YY289 pKa = 11.49LKK291 pKa = 10.15TINKK295 pKa = 8.65WFDD298 pKa = 3.41DD299 pKa = 3.97YY300 pKa = 11.54QGQEE304 pKa = 4.09TIIIDD309 pKa = 4.14DD310 pKa = 4.9LDD312 pKa = 3.71PTEE315 pKa = 4.35AKK317 pKa = 9.82WMGGNLKK324 pKa = 9.93RR325 pKa = 11.84WADD328 pKa = 3.39KK329 pKa = 9.94WPVPVEE335 pKa = 4.3VKK337 pKa = 10.22HH338 pKa = 4.57GHH340 pKa = 4.95IVIRR344 pKa = 11.84PKK346 pKa = 10.62KK347 pKa = 9.9VVVTTQYY354 pKa = 10.96MMEE357 pKa = 4.31QLWDD361 pKa = 3.94DD362 pKa = 3.77PALIEE367 pKa = 4.1ALKK370 pKa = 10.7RR371 pKa = 11.84RR372 pKa = 11.84FKK374 pKa = 10.78IIDD377 pKa = 3.15VDD379 pKa = 4.91LII381 pKa = 4.03
MM1 pKa = 7.45NNNEE5 pKa = 4.12EE6 pKa = 4.21YY7 pKa = 10.59EE8 pKa = 4.01RR9 pKa = 11.84WLNMDD14 pKa = 3.78TFYY17 pKa = 11.67NPLEE21 pKa = 4.04YY22 pKa = 10.51SQTFTPPLSLTTSQQLPPIPMPMTPIEE49 pKa = 4.31GDD51 pKa = 2.99PRR53 pKa = 11.84EE54 pKa = 4.39RR55 pKa = 11.84VGTTAMCGLPRR66 pKa = 11.84AMCAGAPEE74 pKa = 4.17EE75 pKa = 4.46EE76 pKa = 4.42KK77 pKa = 10.94LASAVCALSLEE88 pKa = 4.35DD89 pKa = 4.6LADD92 pKa = 3.61NKK94 pKa = 9.78EE95 pKa = 4.35TFKK98 pKa = 10.97CRR100 pKa = 11.84SIVFTCYY107 pKa = 9.95KK108 pKa = 10.17RR109 pKa = 11.84EE110 pKa = 4.4AIDD113 pKa = 5.93DD114 pKa = 3.93PEE116 pKa = 5.0AWLRR120 pKa = 11.84GLQSKK125 pKa = 8.03CVWVLGQLEE134 pKa = 4.74KK135 pKa = 10.98CPKK138 pKa = 6.67TQRR141 pKa = 11.84IHH143 pKa = 5.39IQGMAYY149 pKa = 10.11NKK151 pKa = 10.42SDD153 pKa = 3.83IAWAMLKK160 pKa = 10.79GEE162 pKa = 4.5STWKK166 pKa = 9.78RR167 pKa = 11.84KK168 pKa = 9.76CIAPIKK174 pKa = 9.85SIEE177 pKa = 4.22YY178 pKa = 7.21CTKK181 pKa = 10.74AKK183 pKa = 10.46SQLAGPWEE191 pKa = 4.62FGDD194 pKa = 3.81RR195 pKa = 11.84PAYY198 pKa = 9.3GQSAKK203 pKa = 9.34TSKK206 pKa = 10.82LNEE209 pKa = 4.27KK210 pKa = 10.71KK211 pKa = 10.69EE212 pKa = 4.11MNATLLTMTPYY223 pKa = 10.35EE224 pKa = 4.53AVDD227 pKa = 3.12EE228 pKa = 5.52GYY230 pKa = 10.37ISWRR234 pKa = 11.84DD235 pKa = 3.54YY236 pKa = 11.54EE237 pKa = 4.24KK238 pKa = 11.25LKK240 pKa = 11.0LFQTTYY246 pKa = 11.07KK247 pKa = 10.19ADD249 pKa = 3.42KK250 pKa = 10.45KK251 pKa = 10.38IAEE254 pKa = 4.19HH255 pKa = 6.47LKK257 pKa = 10.58RR258 pKa = 11.84PIDD261 pKa = 3.26HH262 pKa = 6.82EE263 pKa = 4.39MRR265 pKa = 11.84HH266 pKa = 5.7EE267 pKa = 4.98WIQGKK272 pKa = 8.8SGKK275 pKa = 10.59GKK277 pKa = 4.37TTKK280 pKa = 10.62AILEE284 pKa = 4.47NPDD287 pKa = 4.19HH288 pKa = 6.24YY289 pKa = 11.49LKK291 pKa = 10.15TINKK295 pKa = 8.65WFDD298 pKa = 3.41DD299 pKa = 3.97YY300 pKa = 11.54QGQEE304 pKa = 4.09TIIIDD309 pKa = 4.14DD310 pKa = 4.9LDD312 pKa = 3.71PTEE315 pKa = 4.35AKK317 pKa = 9.82WMGGNLKK324 pKa = 9.93RR325 pKa = 11.84WADD328 pKa = 3.39KK329 pKa = 9.94WPVPVEE335 pKa = 4.3VKK337 pKa = 10.22HH338 pKa = 4.57GHH340 pKa = 4.95IVIRR344 pKa = 11.84PKK346 pKa = 10.62KK347 pKa = 9.9VVVTTQYY354 pKa = 10.96MMEE357 pKa = 4.31QLWDD361 pKa = 3.94DD362 pKa = 3.77PALIEE367 pKa = 4.1ALKK370 pKa = 10.7RR371 pKa = 11.84RR372 pKa = 11.84FKK374 pKa = 10.78IIDD377 pKa = 3.15VDD379 pKa = 4.91LII381 pKa = 4.03
Molecular weight: 43.97 kDa
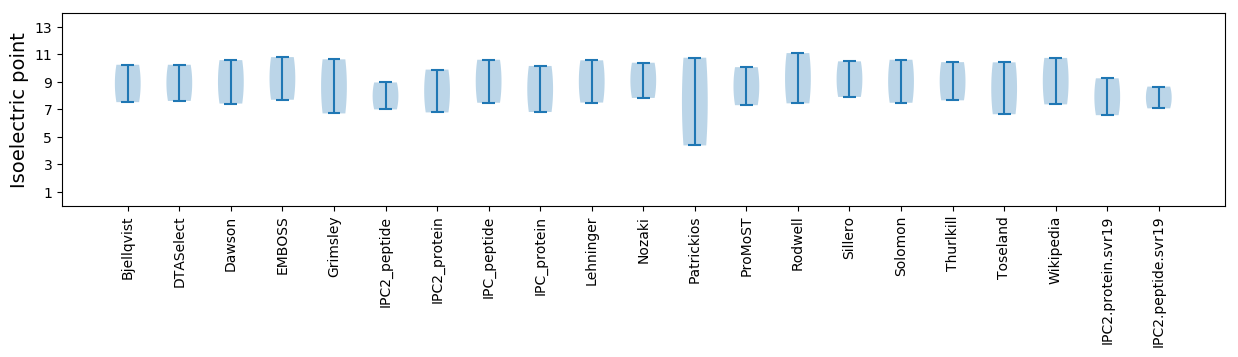
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GAB3|A0A126GAB3_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-33 OX=1685761 PE=4 SV=1
MM1 pKa = 7.11VFYY4 pKa = 10.67GRR6 pKa = 11.84TGGLVGLLSRR16 pKa = 11.84GFGGRR21 pKa = 11.84TGFTKK26 pKa = 10.04PGKK29 pKa = 9.87RR30 pKa = 11.84RR31 pKa = 11.84GSFKK35 pKa = 9.85KK36 pKa = 8.26TFRR39 pKa = 11.84RR40 pKa = 11.84KK41 pKa = 8.39FRR43 pKa = 11.84KK44 pKa = 8.12MRR46 pKa = 11.84YY47 pKa = 8.38RR48 pKa = 11.84KK49 pKa = 9.56KK50 pKa = 10.35RR51 pKa = 11.84YY52 pKa = 7.71NKK54 pKa = 10.14KK55 pKa = 10.34SIGEE59 pKa = 4.01IKK61 pKa = 10.61YY62 pKa = 10.09SPCHH66 pKa = 6.2NGLALTPGQIASTDD80 pKa = 3.49HH81 pKa = 6.79GPATLLQDD89 pKa = 4.07CWPTAGTGVQNVIGRR104 pKa = 11.84QLYY107 pKa = 8.93IRR109 pKa = 11.84KK110 pKa = 9.5LIITLTVANTKK121 pKa = 9.35TNNQQSPMRR130 pKa = 11.84FLLLKK135 pKa = 10.27SKK137 pKa = 10.68KK138 pKa = 9.93FLGGIKK144 pKa = 10.21NNAAFTHH151 pKa = 6.46SSTDD155 pKa = 3.29PAVFPNLFTSTLAYY169 pKa = 9.95SAIPIAQGLGKK180 pKa = 9.14TIFDD184 pKa = 4.29KK185 pKa = 11.23KK186 pKa = 9.35LTPPAAFTTQDD197 pKa = 3.18DD198 pKa = 4.13WVDD201 pKa = 3.16QANYY205 pKa = 8.39VRR207 pKa = 11.84SWKK210 pKa = 8.7KK211 pKa = 8.48TIRR214 pKa = 11.84INKK217 pKa = 9.16VYY219 pKa = 10.36EE220 pKa = 3.5KK221 pKa = 10.48DD222 pKa = 3.19INYY225 pKa = 10.04GSNDD229 pKa = 3.55LFNHH233 pKa = 6.51AGYY236 pKa = 10.55YY237 pKa = 10.13LYY239 pKa = 11.14SLLPTSGQFAGTGAVMNLNYY259 pKa = 10.43YY260 pKa = 8.01FTYY263 pKa = 10.64TDD265 pKa = 3.06VV266 pKa = 3.31
MM1 pKa = 7.11VFYY4 pKa = 10.67GRR6 pKa = 11.84TGGLVGLLSRR16 pKa = 11.84GFGGRR21 pKa = 11.84TGFTKK26 pKa = 10.04PGKK29 pKa = 9.87RR30 pKa = 11.84RR31 pKa = 11.84GSFKK35 pKa = 9.85KK36 pKa = 8.26TFRR39 pKa = 11.84RR40 pKa = 11.84KK41 pKa = 8.39FRR43 pKa = 11.84KK44 pKa = 8.12MRR46 pKa = 11.84YY47 pKa = 8.38RR48 pKa = 11.84KK49 pKa = 9.56KK50 pKa = 10.35RR51 pKa = 11.84YY52 pKa = 7.71NKK54 pKa = 10.14KK55 pKa = 10.34SIGEE59 pKa = 4.01IKK61 pKa = 10.61YY62 pKa = 10.09SPCHH66 pKa = 6.2NGLALTPGQIASTDD80 pKa = 3.49HH81 pKa = 6.79GPATLLQDD89 pKa = 4.07CWPTAGTGVQNVIGRR104 pKa = 11.84QLYY107 pKa = 8.93IRR109 pKa = 11.84KK110 pKa = 9.5LIITLTVANTKK121 pKa = 9.35TNNQQSPMRR130 pKa = 11.84FLLLKK135 pKa = 10.27SKK137 pKa = 10.68KK138 pKa = 9.93FLGGIKK144 pKa = 10.21NNAAFTHH151 pKa = 6.46SSTDD155 pKa = 3.29PAVFPNLFTSTLAYY169 pKa = 9.95SAIPIAQGLGKK180 pKa = 9.14TIFDD184 pKa = 4.29KK185 pKa = 11.23KK186 pKa = 9.35LTPPAAFTTQDD197 pKa = 3.18DD198 pKa = 4.13WVDD201 pKa = 3.16QANYY205 pKa = 8.39VRR207 pKa = 11.84SWKK210 pKa = 8.7KK211 pKa = 8.48TIRR214 pKa = 11.84INKK217 pKa = 9.16VYY219 pKa = 10.36EE220 pKa = 3.5KK221 pKa = 10.48DD222 pKa = 3.19INYY225 pKa = 10.04GSNDD229 pKa = 3.55LFNHH233 pKa = 6.51AGYY236 pKa = 10.55YY237 pKa = 10.13LYY239 pKa = 11.14SLLPTSGQFAGTGAVMNLNYY259 pKa = 10.43YY260 pKa = 8.01FTYY263 pKa = 10.64TDD265 pKa = 3.06VV266 pKa = 3.31
Molecular weight: 29.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
647 |
266 |
381 |
323.5 |
36.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.801 ± 0.256 | 1.7 ± 0.592 |
5.1 ± 0.837 | 5.1 ± 2.713 |
3.709 ± 1.438 | 6.955 ± 1.759 |
1.7 ± 0.123 | 6.028 ± 0.477 |
9.583 ± 0.349 | 8.192 ± 0.284 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.782 ± 0.797 | 4.328 ± 1.053 |
5.564 ± 0.657 | 3.864 ± 0.065 |
5.1 ± 0.57 | 4.637 ± 0.625 |
8.037 ± 1.084 | 3.864 ± 0.169 |
2.473 ± 0.839 | 4.482 ± 0.722 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
