
Brome mosaic virus (BMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Bromovirus
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
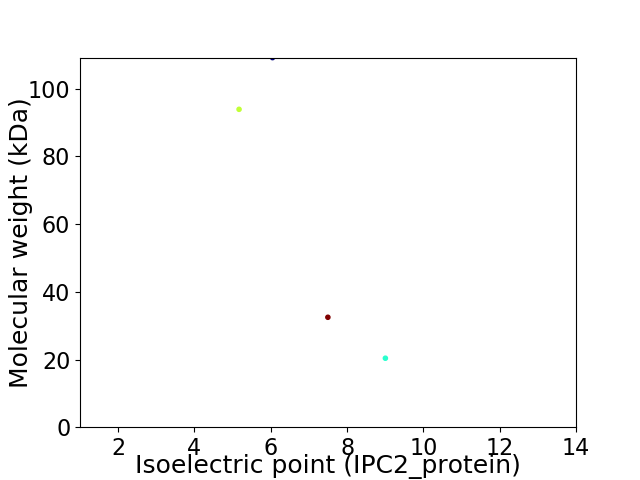
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P03602|CAPSD_BMV Capsid protein OS=Brome mosaic virus OX=12302 GN=ORF3b PE=1 SV=1
MM1 pKa = 7.84SSKK4 pKa = 8.82TWDD7 pKa = 3.48DD8 pKa = 3.54DD9 pKa = 3.75FVRR12 pKa = 11.84QVPSFQWIIDD22 pKa = 3.48QSLEE26 pKa = 4.29DD27 pKa = 3.89EE28 pKa = 4.67VEE30 pKa = 3.99AASLQVQEE38 pKa = 4.79PADD41 pKa = 3.71GVAIDD46 pKa = 4.57GSLASFKK53 pKa = 10.84LAIAPLEE60 pKa = 4.11IGGVFDD66 pKa = 4.38PPFDD70 pKa = 3.66RR71 pKa = 11.84VRR73 pKa = 11.84WGSICDD79 pKa = 3.54TVQQMVQQFTDD90 pKa = 3.43RR91 pKa = 11.84PLIPQAEE98 pKa = 4.68MARR101 pKa = 11.84MLYY104 pKa = 10.58LDD106 pKa = 3.75IPGSFVLEE114 pKa = 4.3DD115 pKa = 5.44EE116 pKa = 4.4IDD118 pKa = 3.22DD119 pKa = 4.41WYY121 pKa = 10.61PEE123 pKa = 4.19DD124 pKa = 3.63TSDD127 pKa = 4.16GYY129 pKa = 11.5GVSFAADD136 pKa = 3.54EE137 pKa = 4.32DD138 pKa = 4.23HH139 pKa = 7.85ASDD142 pKa = 5.12LKK144 pKa = 11.1LASDD148 pKa = 4.15SSNCEE153 pKa = 3.48IEE155 pKa = 4.34EE156 pKa = 3.99VRR158 pKa = 11.84VTGDD162 pKa = 3.25TPKK165 pKa = 10.81EE166 pKa = 3.76LTLGDD171 pKa = 3.74RR172 pKa = 11.84YY173 pKa = 10.38MGIDD177 pKa = 4.16EE178 pKa = 4.86EE179 pKa = 4.57FQTTNTDD186 pKa = 2.9YY187 pKa = 11.31DD188 pKa = 3.39ITLQIMNPIEE198 pKa = 4.09HH199 pKa = 6.3RR200 pKa = 11.84VSRR203 pKa = 11.84VIDD206 pKa = 3.6THH208 pKa = 6.04CHH210 pKa = 6.27PDD212 pKa = 3.36NPDD215 pKa = 2.87ISTGPIYY222 pKa = 9.77MEE224 pKa = 3.97RR225 pKa = 11.84VSLARR230 pKa = 11.84TEE232 pKa = 4.27ATSHH236 pKa = 6.62SILPTHH242 pKa = 7.35AYY244 pKa = 10.36FDD246 pKa = 4.38DD247 pKa = 4.87SYY249 pKa = 11.63HH250 pKa = 5.86QALVEE255 pKa = 4.22NGDD258 pKa = 3.4YY259 pKa = 11.59SMDD262 pKa = 3.65FDD264 pKa = 6.27RR265 pKa = 11.84IRR267 pKa = 11.84LKK269 pKa = 10.82QSDD272 pKa = 4.15VDD274 pKa = 3.6WYY276 pKa = 11.02RR277 pKa = 11.84DD278 pKa = 3.3PDD280 pKa = 3.78KK281 pKa = 11.4YY282 pKa = 10.76FQPKK286 pKa = 8.96MNIGSAQRR294 pKa = 11.84RR295 pKa = 11.84VGTQKK300 pKa = 10.68EE301 pKa = 4.29VLTALKK307 pKa = 10.1KK308 pKa = 10.58RR309 pKa = 11.84NADD312 pKa = 3.46VPEE315 pKa = 4.39MGDD318 pKa = 3.95AINMKK323 pKa = 9.12DD324 pKa = 3.25TAKK327 pKa = 10.79AIAKK331 pKa = 9.48RR332 pKa = 11.84FRR334 pKa = 11.84STFLNVDD341 pKa = 3.39GEE343 pKa = 4.57DD344 pKa = 3.82CLRR347 pKa = 11.84ASMDD351 pKa = 3.27VMTKK355 pKa = 10.22CLEE358 pKa = 4.03YY359 pKa = 9.85HH360 pKa = 6.25KK361 pKa = 10.73KK362 pKa = 8.54WGKK365 pKa = 10.03HH366 pKa = 4.31MDD368 pKa = 3.53LQGVNVAAEE377 pKa = 4.21TDD379 pKa = 3.45LCRR382 pKa = 11.84YY383 pKa = 7.52QHH385 pKa = 5.81MLKK388 pKa = 10.29SDD390 pKa = 3.81VKK392 pKa = 10.66PVVTDD397 pKa = 3.44TLHH400 pKa = 7.33LEE402 pKa = 4.03RR403 pKa = 11.84AVAATITFHH412 pKa = 7.15SKK414 pKa = 10.29GVTSNFSPFFTACFEE429 pKa = 4.28KK430 pKa = 10.91LSLALKK436 pKa = 10.44SRR438 pKa = 11.84FIVPIGKK445 pKa = 9.47ISSLEE450 pKa = 4.01LKK452 pKa = 10.3NVRR455 pKa = 11.84LNNRR459 pKa = 11.84YY460 pKa = 8.9FLEE463 pKa = 5.12ADD465 pKa = 3.35LSKK468 pKa = 10.63FDD470 pKa = 3.7KK471 pKa = 11.18SQGEE475 pKa = 3.98LHH477 pKa = 7.06LEE479 pKa = 4.13FQRR482 pKa = 11.84EE483 pKa = 3.73ILLALGFPAPLTNWWSDD500 pKa = 4.21FHH502 pKa = 7.43RR503 pKa = 11.84DD504 pKa = 3.28SYY506 pKa = 11.67LSDD509 pKa = 3.12PHH511 pKa = 8.2AKK513 pKa = 10.16VGMSVSFQRR522 pKa = 11.84RR523 pKa = 11.84TGDD526 pKa = 2.78AFTYY530 pKa = 10.33FGNTLVTMAMIAYY543 pKa = 9.86ASDD546 pKa = 5.5LSDD549 pKa = 3.87CDD551 pKa = 3.8CAIFSGDD558 pKa = 3.74DD559 pKa = 3.21SLIISKK565 pKa = 9.29VKK567 pKa = 10.21PVLDD571 pKa = 3.36TDD573 pKa = 3.79MFTSLFNMEE582 pKa = 4.84IKK584 pKa = 11.01VMDD587 pKa = 4.23PSVPYY592 pKa = 9.97VCSKK596 pKa = 10.59FLVEE600 pKa = 4.3TEE602 pKa = 3.89MGNLVSVPDD611 pKa = 4.47PLRR614 pKa = 11.84EE615 pKa = 3.79IQRR618 pKa = 11.84LAKK621 pKa = 10.23RR622 pKa = 11.84KK623 pKa = 9.27ILRR626 pKa = 11.84DD627 pKa = 3.46EE628 pKa = 4.15QMLRR632 pKa = 11.84AHH634 pKa = 6.69FVSFCDD640 pKa = 2.86RR641 pKa = 11.84MKK643 pKa = 10.97FINQLDD649 pKa = 3.82EE650 pKa = 5.16KK651 pKa = 10.47MITTLCHH658 pKa = 5.93FVYY661 pKa = 10.47LKK663 pKa = 10.61YY664 pKa = 10.72GKK666 pKa = 9.08EE667 pKa = 3.9KK668 pKa = 9.37PWIFEE673 pKa = 3.91EE674 pKa = 4.57VRR676 pKa = 11.84AALAAFSLYY685 pKa = 10.71SEE687 pKa = 4.15NFLRR691 pKa = 11.84FSDD694 pKa = 4.81CYY696 pKa = 8.1CTEE699 pKa = 4.83GIRR702 pKa = 11.84VYY704 pKa = 11.13QMSDD708 pKa = 3.25PVCKK712 pKa = 10.67FKK714 pKa = 10.62RR715 pKa = 11.84TTEE718 pKa = 3.83EE719 pKa = 4.05RR720 pKa = 11.84KK721 pKa = 8.3TDD723 pKa = 3.54GDD725 pKa = 3.7WFHH728 pKa = 6.22NWKK731 pKa = 9.94NPKK734 pKa = 9.91FPGVLDD740 pKa = 3.48KK741 pKa = 11.06VYY743 pKa = 9.43RR744 pKa = 11.84TIGIYY749 pKa = 10.59SSDD752 pKa = 3.94CSTKK756 pKa = 9.32EE757 pKa = 3.88LPVKK761 pKa = 10.55RR762 pKa = 11.84IGRR765 pKa = 11.84LHH767 pKa = 6.32EE768 pKa = 4.23ALEE771 pKa = 4.32RR772 pKa = 11.84EE773 pKa = 4.41SLKK776 pKa = 10.75LANDD780 pKa = 3.08RR781 pKa = 11.84RR782 pKa = 11.84TTQRR786 pKa = 11.84LKK788 pKa = 11.14KK789 pKa = 10.45KK790 pKa = 10.11VDD792 pKa = 3.73DD793 pKa = 4.02YY794 pKa = 11.18ATGRR798 pKa = 11.84GGLTSVDD805 pKa = 3.18ALLVKK810 pKa = 8.2SHH812 pKa = 7.24CEE814 pKa = 3.65TFKK817 pKa = 11.13PSDD820 pKa = 3.46LRR822 pKa = 4.93
MM1 pKa = 7.84SSKK4 pKa = 8.82TWDD7 pKa = 3.48DD8 pKa = 3.54DD9 pKa = 3.75FVRR12 pKa = 11.84QVPSFQWIIDD22 pKa = 3.48QSLEE26 pKa = 4.29DD27 pKa = 3.89EE28 pKa = 4.67VEE30 pKa = 3.99AASLQVQEE38 pKa = 4.79PADD41 pKa = 3.71GVAIDD46 pKa = 4.57GSLASFKK53 pKa = 10.84LAIAPLEE60 pKa = 4.11IGGVFDD66 pKa = 4.38PPFDD70 pKa = 3.66RR71 pKa = 11.84VRR73 pKa = 11.84WGSICDD79 pKa = 3.54TVQQMVQQFTDD90 pKa = 3.43RR91 pKa = 11.84PLIPQAEE98 pKa = 4.68MARR101 pKa = 11.84MLYY104 pKa = 10.58LDD106 pKa = 3.75IPGSFVLEE114 pKa = 4.3DD115 pKa = 5.44EE116 pKa = 4.4IDD118 pKa = 3.22DD119 pKa = 4.41WYY121 pKa = 10.61PEE123 pKa = 4.19DD124 pKa = 3.63TSDD127 pKa = 4.16GYY129 pKa = 11.5GVSFAADD136 pKa = 3.54EE137 pKa = 4.32DD138 pKa = 4.23HH139 pKa = 7.85ASDD142 pKa = 5.12LKK144 pKa = 11.1LASDD148 pKa = 4.15SSNCEE153 pKa = 3.48IEE155 pKa = 4.34EE156 pKa = 3.99VRR158 pKa = 11.84VTGDD162 pKa = 3.25TPKK165 pKa = 10.81EE166 pKa = 3.76LTLGDD171 pKa = 3.74RR172 pKa = 11.84YY173 pKa = 10.38MGIDD177 pKa = 4.16EE178 pKa = 4.86EE179 pKa = 4.57FQTTNTDD186 pKa = 2.9YY187 pKa = 11.31DD188 pKa = 3.39ITLQIMNPIEE198 pKa = 4.09HH199 pKa = 6.3RR200 pKa = 11.84VSRR203 pKa = 11.84VIDD206 pKa = 3.6THH208 pKa = 6.04CHH210 pKa = 6.27PDD212 pKa = 3.36NPDD215 pKa = 2.87ISTGPIYY222 pKa = 9.77MEE224 pKa = 3.97RR225 pKa = 11.84VSLARR230 pKa = 11.84TEE232 pKa = 4.27ATSHH236 pKa = 6.62SILPTHH242 pKa = 7.35AYY244 pKa = 10.36FDD246 pKa = 4.38DD247 pKa = 4.87SYY249 pKa = 11.63HH250 pKa = 5.86QALVEE255 pKa = 4.22NGDD258 pKa = 3.4YY259 pKa = 11.59SMDD262 pKa = 3.65FDD264 pKa = 6.27RR265 pKa = 11.84IRR267 pKa = 11.84LKK269 pKa = 10.82QSDD272 pKa = 4.15VDD274 pKa = 3.6WYY276 pKa = 11.02RR277 pKa = 11.84DD278 pKa = 3.3PDD280 pKa = 3.78KK281 pKa = 11.4YY282 pKa = 10.76FQPKK286 pKa = 8.96MNIGSAQRR294 pKa = 11.84RR295 pKa = 11.84VGTQKK300 pKa = 10.68EE301 pKa = 4.29VLTALKK307 pKa = 10.1KK308 pKa = 10.58RR309 pKa = 11.84NADD312 pKa = 3.46VPEE315 pKa = 4.39MGDD318 pKa = 3.95AINMKK323 pKa = 9.12DD324 pKa = 3.25TAKK327 pKa = 10.79AIAKK331 pKa = 9.48RR332 pKa = 11.84FRR334 pKa = 11.84STFLNVDD341 pKa = 3.39GEE343 pKa = 4.57DD344 pKa = 3.82CLRR347 pKa = 11.84ASMDD351 pKa = 3.27VMTKK355 pKa = 10.22CLEE358 pKa = 4.03YY359 pKa = 9.85HH360 pKa = 6.25KK361 pKa = 10.73KK362 pKa = 8.54WGKK365 pKa = 10.03HH366 pKa = 4.31MDD368 pKa = 3.53LQGVNVAAEE377 pKa = 4.21TDD379 pKa = 3.45LCRR382 pKa = 11.84YY383 pKa = 7.52QHH385 pKa = 5.81MLKK388 pKa = 10.29SDD390 pKa = 3.81VKK392 pKa = 10.66PVVTDD397 pKa = 3.44TLHH400 pKa = 7.33LEE402 pKa = 4.03RR403 pKa = 11.84AVAATITFHH412 pKa = 7.15SKK414 pKa = 10.29GVTSNFSPFFTACFEE429 pKa = 4.28KK430 pKa = 10.91LSLALKK436 pKa = 10.44SRR438 pKa = 11.84FIVPIGKK445 pKa = 9.47ISSLEE450 pKa = 4.01LKK452 pKa = 10.3NVRR455 pKa = 11.84LNNRR459 pKa = 11.84YY460 pKa = 8.9FLEE463 pKa = 5.12ADD465 pKa = 3.35LSKK468 pKa = 10.63FDD470 pKa = 3.7KK471 pKa = 11.18SQGEE475 pKa = 3.98LHH477 pKa = 7.06LEE479 pKa = 4.13FQRR482 pKa = 11.84EE483 pKa = 3.73ILLALGFPAPLTNWWSDD500 pKa = 4.21FHH502 pKa = 7.43RR503 pKa = 11.84DD504 pKa = 3.28SYY506 pKa = 11.67LSDD509 pKa = 3.12PHH511 pKa = 8.2AKK513 pKa = 10.16VGMSVSFQRR522 pKa = 11.84RR523 pKa = 11.84TGDD526 pKa = 2.78AFTYY530 pKa = 10.33FGNTLVTMAMIAYY543 pKa = 9.86ASDD546 pKa = 5.5LSDD549 pKa = 3.87CDD551 pKa = 3.8CAIFSGDD558 pKa = 3.74DD559 pKa = 3.21SLIISKK565 pKa = 9.29VKK567 pKa = 10.21PVLDD571 pKa = 3.36TDD573 pKa = 3.79MFTSLFNMEE582 pKa = 4.84IKK584 pKa = 11.01VMDD587 pKa = 4.23PSVPYY592 pKa = 9.97VCSKK596 pKa = 10.59FLVEE600 pKa = 4.3TEE602 pKa = 3.89MGNLVSVPDD611 pKa = 4.47PLRR614 pKa = 11.84EE615 pKa = 3.79IQRR618 pKa = 11.84LAKK621 pKa = 10.23RR622 pKa = 11.84KK623 pKa = 9.27ILRR626 pKa = 11.84DD627 pKa = 3.46EE628 pKa = 4.15QMLRR632 pKa = 11.84AHH634 pKa = 6.69FVSFCDD640 pKa = 2.86RR641 pKa = 11.84MKK643 pKa = 10.97FINQLDD649 pKa = 3.82EE650 pKa = 5.16KK651 pKa = 10.47MITTLCHH658 pKa = 5.93FVYY661 pKa = 10.47LKK663 pKa = 10.61YY664 pKa = 10.72GKK666 pKa = 9.08EE667 pKa = 3.9KK668 pKa = 9.37PWIFEE673 pKa = 3.91EE674 pKa = 4.57VRR676 pKa = 11.84AALAAFSLYY685 pKa = 10.71SEE687 pKa = 4.15NFLRR691 pKa = 11.84FSDD694 pKa = 4.81CYY696 pKa = 8.1CTEE699 pKa = 4.83GIRR702 pKa = 11.84VYY704 pKa = 11.13QMSDD708 pKa = 3.25PVCKK712 pKa = 10.67FKK714 pKa = 10.62RR715 pKa = 11.84TTEE718 pKa = 3.83EE719 pKa = 4.05RR720 pKa = 11.84KK721 pKa = 8.3TDD723 pKa = 3.54GDD725 pKa = 3.7WFHH728 pKa = 6.22NWKK731 pKa = 9.94NPKK734 pKa = 9.91FPGVLDD740 pKa = 3.48KK741 pKa = 11.06VYY743 pKa = 9.43RR744 pKa = 11.84TIGIYY749 pKa = 10.59SSDD752 pKa = 3.94CSTKK756 pKa = 9.32EE757 pKa = 3.88LPVKK761 pKa = 10.55RR762 pKa = 11.84IGRR765 pKa = 11.84LHH767 pKa = 6.32EE768 pKa = 4.23ALEE771 pKa = 4.32RR772 pKa = 11.84EE773 pKa = 4.41SLKK776 pKa = 10.75LANDD780 pKa = 3.08RR781 pKa = 11.84RR782 pKa = 11.84TTQRR786 pKa = 11.84LKK788 pKa = 11.14KK789 pKa = 10.45KK790 pKa = 10.11VDD792 pKa = 3.73DD793 pKa = 4.02YY794 pKa = 11.18ATGRR798 pKa = 11.84GGLTSVDD805 pKa = 3.18ALLVKK810 pKa = 8.2SHH812 pKa = 7.24CEE814 pKa = 3.65TFKK817 pKa = 11.13PSDD820 pKa = 3.46LRR822 pKa = 4.93
Molecular weight: 93.93 kDa
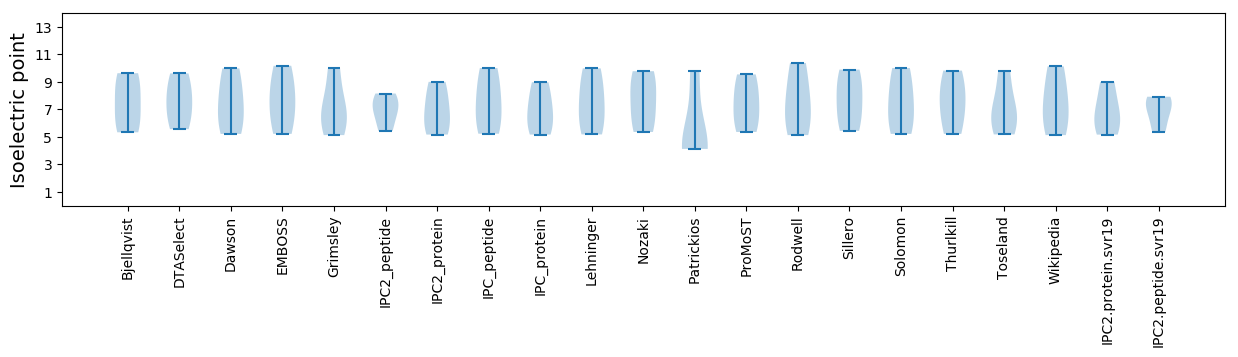
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03603|MVP_BMV Movement protein OS=Brome mosaic virus OX=12302 GN=ORF3a PE=1 SV=1
MM1 pKa = 6.91STSGTGKK8 pKa = 7.38MTRR11 pKa = 11.84AQRR14 pKa = 11.84RR15 pKa = 11.84AAARR19 pKa = 11.84RR20 pKa = 11.84NRR22 pKa = 11.84WTARR26 pKa = 11.84VQPVIVEE33 pKa = 4.05PLAAGQGKK41 pKa = 9.95AIKK44 pKa = 10.19AIAGYY49 pKa = 10.07SISKK53 pKa = 9.61WEE55 pKa = 4.16ASSDD59 pKa = 3.85AITAKK64 pKa = 9.48ATNAMSITLPHH75 pKa = 6.47EE76 pKa = 4.38LSSEE80 pKa = 4.06KK81 pKa = 10.79NKK83 pKa = 9.76EE84 pKa = 4.11LKK86 pKa = 10.11VGRR89 pKa = 11.84VLLWLGLLPSVAGRR103 pKa = 11.84IKK105 pKa = 10.67ACVAEE110 pKa = 4.39KK111 pKa = 10.23QAQAEE116 pKa = 4.2AAFQVALAVADD127 pKa = 3.7SSKK130 pKa = 10.93EE131 pKa = 3.86VVAAMYY137 pKa = 9.95TDD139 pKa = 4.01AFRR142 pKa = 11.84GATLGDD148 pKa = 3.96LLNLQIYY155 pKa = 9.3LYY157 pKa = 10.83ASEE160 pKa = 4.94AVPAKK165 pKa = 10.38AVVVHH170 pKa = 6.37LEE172 pKa = 4.18VEE174 pKa = 4.59HH175 pKa = 6.17VRR177 pKa = 11.84PTFDD181 pKa = 5.07DD182 pKa = 4.32FFTPVYY188 pKa = 10.25RR189 pKa = 5.11
MM1 pKa = 6.91STSGTGKK8 pKa = 7.38MTRR11 pKa = 11.84AQRR14 pKa = 11.84RR15 pKa = 11.84AAARR19 pKa = 11.84RR20 pKa = 11.84NRR22 pKa = 11.84WTARR26 pKa = 11.84VQPVIVEE33 pKa = 4.05PLAAGQGKK41 pKa = 9.95AIKK44 pKa = 10.19AIAGYY49 pKa = 10.07SISKK53 pKa = 9.61WEE55 pKa = 4.16ASSDD59 pKa = 3.85AITAKK64 pKa = 9.48ATNAMSITLPHH75 pKa = 6.47EE76 pKa = 4.38LSSEE80 pKa = 4.06KK81 pKa = 10.79NKK83 pKa = 9.76EE84 pKa = 4.11LKK86 pKa = 10.11VGRR89 pKa = 11.84VLLWLGLLPSVAGRR103 pKa = 11.84IKK105 pKa = 10.67ACVAEE110 pKa = 4.39KK111 pKa = 10.23QAQAEE116 pKa = 4.2AAFQVALAVADD127 pKa = 3.7SSKK130 pKa = 10.93EE131 pKa = 3.86VVAAMYY137 pKa = 9.95TDD139 pKa = 4.01AFRR142 pKa = 11.84GATLGDD148 pKa = 3.96LLNLQIYY155 pKa = 9.3LYY157 pKa = 10.83ASEE160 pKa = 4.94AVPAKK165 pKa = 10.38AVVVHH170 pKa = 6.37LEE172 pKa = 4.18VEE174 pKa = 4.59HH175 pKa = 6.17VRR177 pKa = 11.84PTFDD181 pKa = 5.07DD182 pKa = 4.32FFTPVYY188 pKa = 10.25RR189 pKa = 5.11
Molecular weight: 20.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2275 |
189 |
961 |
568.8 |
64.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.956 ± 1.434 | 2.33 ± 0.384 |
7.604 ± 0.99 | 6.022 ± 0.216 |
4.22 ± 0.559 | 5.451 ± 0.517 |
2.725 ± 0.468 | 4.791 ± 0.111 |
6.418 ± 0.068 | 8.484 ± 0.134 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.374 ± 0.344 | 3.121 ± 0.229 |
3.736 ± 0.415 | 3.209 ± 0.106 |
6.11 ± 0.319 | 8.132 ± 0.826 |
5.451 ± 0.246 | 7.297 ± 0.414 |
1.582 ± 0.199 | 2.989 ± 0.082 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
