
Burkholderiales bacterium GJ-E10
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; unclassified Burkholderiales
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
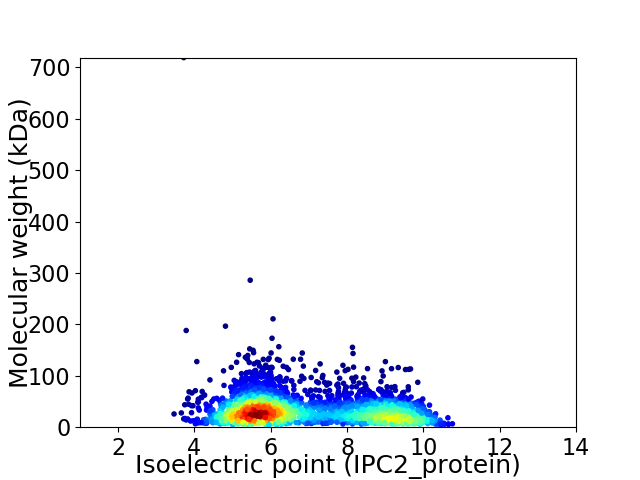
Virtual 2D-PAGE plot for 2955 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1H0I7|A0A0A1H0I7_9BURK Uncharacterized protein OS=Burkholderiales bacterium GJ-E10 OX=1469502 GN=E1O_06300 PE=4 SV=1
MM1 pKa = 7.91TITQATQNAAINWQSFNIGSNQSVTFVQPNASAVALNRR39 pKa = 11.84VLGPDD44 pKa = 3.91PSNIWGTLDD53 pKa = 3.64ANGKK57 pKa = 9.41VFLINPNGILFGKK70 pKa = 9.54GASINVGGLVASTLGLSDD88 pKa = 4.33RR89 pKa = 11.84DD90 pKa = 3.82FLAGNYY96 pKa = 9.1RR97 pKa = 11.84FSGASAAAVQNQGSITADD115 pKa = 2.97GGYY118 pKa = 10.08VALLGANVSNQGVIAARR135 pKa = 11.84LGTVALAGGNAVTLDD150 pKa = 3.51VAGDD154 pKa = 3.83GLLSVAVDD162 pKa = 3.39QGAVHH167 pKa = 7.22ALVQNGGLIQADD179 pKa = 3.92GGTVLLTTQAAGNLLSTAVNDD200 pKa = 3.67SGIIEE205 pKa = 4.4AQTVEE210 pKa = 4.32SHH212 pKa = 6.32NGTIRR217 pKa = 11.84LLGDD221 pKa = 3.53MQSGTVTVSGLLDD234 pKa = 3.77ASAPNGGNGGNIEE247 pKa = 4.14TSAAMLDD254 pKa = 3.5ISPTVAITTAAPSGTAGTWLLDD276 pKa = 3.92PYY278 pKa = 11.25NVTISTAADD287 pKa = 3.54SNTSAFTATASGANINTTTLQNALATTSVTVTTGSSGTEE326 pKa = 3.67NGDD329 pKa = 3.26ITVSAPISWSANTNLTLAAAGSIYY353 pKa = 10.79LNAGIANSGGGSIEE367 pKa = 3.95AKK369 pKa = 10.05AAGNIIQAGVGITTQGGNVVYY390 pKa = 9.56WANDD394 pKa = 3.39AGNGGYY400 pKa = 9.56VEE402 pKa = 4.43LQSGSSITTNGGGLWIGGGSGTAQWTPYY430 pKa = 10.2AGASAITVGNGYY442 pKa = 10.61AVGNATLTASGSTNFEE458 pKa = 4.0SGVIVAGPNSGAPTVSLDD476 pKa = 3.98TISSGSTDD484 pKa = 2.87GSISINGATPATAISGDD501 pKa = 3.44NVQGVTIYY509 pKa = 10.4GANVSSGRR517 pKa = 11.84GTLTINGQGSTTAGSPGPVQAIEE540 pKa = 4.59FGNSTLSSTSGAISLTGDD558 pKa = 3.0ASGGSGSNQFLGIHH572 pKa = 6.71WDD574 pKa = 3.46PSGTVASTTGAVNITGYY591 pKa = 10.97SDD593 pKa = 4.24INGQPWSSQDD603 pKa = 2.98VGLAMQGTLGGAGEE617 pKa = 4.79HH618 pKa = 6.85GNITLQTDD626 pKa = 3.61VFLFDD631 pKa = 3.99SGTDD635 pKa = 3.63LVQSTGGLTILPKK648 pKa = 10.5AGEE651 pKa = 4.22GLVVSASTPSTNSAPEE667 pKa = 3.77TLMPIWFNTSGTYY680 pKa = 8.77FVPGFSGITLGSSGSTGAVTVSSPLSVADD709 pKa = 3.96PLTINGSTVTVGANLTNTALNSAILVQATGDD740 pKa = 3.37ITQTASAAVTTNGGSVTYY758 pKa = 10.29DD759 pKa = 3.21SNSANLGSGNIWLQGTGTSGASINTNGGNITLSGGSNVATGYY801 pKa = 9.05ATGDD805 pKa = 3.18AALNGNGVSLDD816 pKa = 3.73TVNLFSGGGNIVIRR830 pKa = 11.84GKK832 pKa = 10.81SSTTNPSFSTSEE844 pKa = 4.02CSPCNTNDD852 pKa = 4.26GIHH855 pKa = 6.31MYY857 pKa = 10.81GSDD860 pKa = 4.15VINAGSGTIDD870 pKa = 3.58LEE872 pKa = 4.62GVAVGNSSSQYY883 pKa = 9.81SNGIEE888 pKa = 4.06TSITGYY894 pKa = 8.63TRR896 pKa = 11.84ILSSATNATAINLVGDD912 pKa = 4.46ASASTSSSDD921 pKa = 2.94EE922 pKa = 3.72WGVFLWGGNNFGIVVAATGTGGGVSISGKK951 pKa = 10.2GGNFTGKK958 pKa = 10.28AGGTHH963 pKa = 6.07TEE965 pKa = 3.71PNAYY969 pKa = 9.59VLAASGPISLTGTAGALSTTYY990 pKa = 11.37GDD992 pKa = 3.1VDD994 pKa = 4.42LNSTVGFVSTLPSGFGIASPVTSSSSNISITADD1027 pKa = 3.15SFKK1030 pKa = 10.41TDD1032 pKa = 3.22VVFGSGTFTSSAVQSSGTLSIAPRR1056 pKa = 11.84SPGKK1060 pKa = 10.38ALAVQTTPPSGTPLWINPTTMFGSSGLFKK1089 pKa = 10.46TGFSSFVFGSASTGTVTLDD1108 pKa = 2.83NYY1110 pKa = 10.67TFDD1113 pKa = 3.89NVTTVDD1119 pKa = 3.22TGANAILGTISIPNHH1134 pKa = 5.99ALTVNITGSGSVTDD1148 pKa = 3.57TGVIDD1153 pKa = 4.14VSSLALDD1160 pKa = 4.04AASSAVTLDD1169 pKa = 3.27NTGNAIGTLAADD1181 pKa = 3.9VASLSLDD1188 pKa = 3.22NATGLTVGSVAALNGVSATGGIDD1211 pKa = 3.01IEE1213 pKa = 4.57TATGSLTVTQNIATSDD1229 pKa = 3.5TGSAAIVLDD1238 pKa = 4.18AGNATAAGIATGGDD1252 pKa = 3.03IVLGGTPSPSISVGSGGKK1270 pKa = 9.6AVLYY1274 pKa = 8.73TGSVSGSTGIGALVGSGNSRR1294 pKa = 11.84YY1295 pKa = 9.97DD1296 pKa = 3.29SSKK1299 pKa = 7.27TTPGFTVALGSSGMFAVYY1317 pKa = 10.12RR1318 pKa = 11.84EE1319 pKa = 3.97QPILGYY1325 pKa = 8.24TAGSATITYY1334 pKa = 8.95GGSVPMSSLTPTYY1347 pKa = 10.53TSGLVNGDD1355 pKa = 3.45ALAQVVSSVPSASYY1369 pKa = 10.15TSSLSSGGYY1378 pKa = 8.96QNAGIYY1384 pKa = 9.92AVSASGGVTGFGYY1397 pKa = 10.58GFQYY1401 pKa = 10.62TGGTLTVNAKK1411 pKa = 10.32ALTITASAQSTTYY1424 pKa = 9.15GTALALGTSQFTPSGLVNGDD1444 pKa = 3.23SVAGVTLTQGGNATVPVTQAAGTYY1468 pKa = 8.31TGSTNGILASGATGTGLSNYY1488 pKa = 7.82TISYY1492 pKa = 9.98VPGTLTIDD1500 pKa = 3.82PKK1502 pKa = 11.25ALTVSGEE1509 pKa = 3.71AALNKK1514 pKa = 9.88VYY1516 pKa = 10.74DD1517 pKa = 4.1GSTTATLTGGTLPGLINGDD1536 pKa = 3.91SVTLTQAGTFASKK1549 pKa = 10.9NVGTNITVTASDD1561 pKa = 4.52SISGPSAGNYY1571 pKa = 8.76TLMQPTGLTANITQLASVTWIGGASGNWFDD1601 pKa = 3.78PANWAGGAVPDD1612 pKa = 4.64LSNVANVVIPQGVTVTFDD1630 pKa = 3.19SANLVAPAQGGPVNLASVGSAGNLTMAAGTLNVSSGVRR1668 pKa = 11.84LNTLAQTGGTVDD1680 pKa = 3.14SNGPVAVDD1688 pKa = 3.39VLTQSGGTLGGAGSVTASSLMQSGGSIVVGGNVTAGTLTQTAGSIDD1734 pKa = 3.28NGGNVGAVALAQSGGQIANRR1754 pKa = 11.84GGLDD1758 pKa = 3.26VTQSFSQSAPGSIAVGGSIAITQAAGNLSFVSLAGNDD1795 pKa = 2.91IDD1797 pKa = 5.75LGANSGAVNLGALDD1811 pKa = 3.77AAGSLTVNASGNISQQPGSVIRR1833 pKa = 11.84VATNVAMNSTAGAVTVSTAGNSFGSGLDD1861 pKa = 3.45SASPAQAAQSSVAADD1876 pKa = 3.43VEE1878 pKa = 4.73SGIEE1882 pKa = 3.95FTPEE1886 pKa = 3.22AGNLVEE1892 pKa = 4.78PALHH1896 pKa = 6.2TEE1898 pKa = 4.77PGQGMGTAGGSSEE1911 pKa = 3.95PAVAQEE1917 pKa = 4.22SGGAAVQPLAGIRR1930 pKa = 11.84LTVLDD1935 pKa = 3.96GGVRR1939 pKa = 11.84LPEE1942 pKa = 3.95LRR1944 pKa = 4.39
MM1 pKa = 7.91TITQATQNAAINWQSFNIGSNQSVTFVQPNASAVALNRR39 pKa = 11.84VLGPDD44 pKa = 3.91PSNIWGTLDD53 pKa = 3.64ANGKK57 pKa = 9.41VFLINPNGILFGKK70 pKa = 9.54GASINVGGLVASTLGLSDD88 pKa = 4.33RR89 pKa = 11.84DD90 pKa = 3.82FLAGNYY96 pKa = 9.1RR97 pKa = 11.84FSGASAAAVQNQGSITADD115 pKa = 2.97GGYY118 pKa = 10.08VALLGANVSNQGVIAARR135 pKa = 11.84LGTVALAGGNAVTLDD150 pKa = 3.51VAGDD154 pKa = 3.83GLLSVAVDD162 pKa = 3.39QGAVHH167 pKa = 7.22ALVQNGGLIQADD179 pKa = 3.92GGTVLLTTQAAGNLLSTAVNDD200 pKa = 3.67SGIIEE205 pKa = 4.4AQTVEE210 pKa = 4.32SHH212 pKa = 6.32NGTIRR217 pKa = 11.84LLGDD221 pKa = 3.53MQSGTVTVSGLLDD234 pKa = 3.77ASAPNGGNGGNIEE247 pKa = 4.14TSAAMLDD254 pKa = 3.5ISPTVAITTAAPSGTAGTWLLDD276 pKa = 3.92PYY278 pKa = 11.25NVTISTAADD287 pKa = 3.54SNTSAFTATASGANINTTTLQNALATTSVTVTTGSSGTEE326 pKa = 3.67NGDD329 pKa = 3.26ITVSAPISWSANTNLTLAAAGSIYY353 pKa = 10.79LNAGIANSGGGSIEE367 pKa = 3.95AKK369 pKa = 10.05AAGNIIQAGVGITTQGGNVVYY390 pKa = 9.56WANDD394 pKa = 3.39AGNGGYY400 pKa = 9.56VEE402 pKa = 4.43LQSGSSITTNGGGLWIGGGSGTAQWTPYY430 pKa = 10.2AGASAITVGNGYY442 pKa = 10.61AVGNATLTASGSTNFEE458 pKa = 4.0SGVIVAGPNSGAPTVSLDD476 pKa = 3.98TISSGSTDD484 pKa = 2.87GSISINGATPATAISGDD501 pKa = 3.44NVQGVTIYY509 pKa = 10.4GANVSSGRR517 pKa = 11.84GTLTINGQGSTTAGSPGPVQAIEE540 pKa = 4.59FGNSTLSSTSGAISLTGDD558 pKa = 3.0ASGGSGSNQFLGIHH572 pKa = 6.71WDD574 pKa = 3.46PSGTVASTTGAVNITGYY591 pKa = 10.97SDD593 pKa = 4.24INGQPWSSQDD603 pKa = 2.98VGLAMQGTLGGAGEE617 pKa = 4.79HH618 pKa = 6.85GNITLQTDD626 pKa = 3.61VFLFDD631 pKa = 3.99SGTDD635 pKa = 3.63LVQSTGGLTILPKK648 pKa = 10.5AGEE651 pKa = 4.22GLVVSASTPSTNSAPEE667 pKa = 3.77TLMPIWFNTSGTYY680 pKa = 8.77FVPGFSGITLGSSGSTGAVTVSSPLSVADD709 pKa = 3.96PLTINGSTVTVGANLTNTALNSAILVQATGDD740 pKa = 3.37ITQTASAAVTTNGGSVTYY758 pKa = 10.29DD759 pKa = 3.21SNSANLGSGNIWLQGTGTSGASINTNGGNITLSGGSNVATGYY801 pKa = 9.05ATGDD805 pKa = 3.18AALNGNGVSLDD816 pKa = 3.73TVNLFSGGGNIVIRR830 pKa = 11.84GKK832 pKa = 10.81SSTTNPSFSTSEE844 pKa = 4.02CSPCNTNDD852 pKa = 4.26GIHH855 pKa = 6.31MYY857 pKa = 10.81GSDD860 pKa = 4.15VINAGSGTIDD870 pKa = 3.58LEE872 pKa = 4.62GVAVGNSSSQYY883 pKa = 9.81SNGIEE888 pKa = 4.06TSITGYY894 pKa = 8.63TRR896 pKa = 11.84ILSSATNATAINLVGDD912 pKa = 4.46ASASTSSSDD921 pKa = 2.94EE922 pKa = 3.72WGVFLWGGNNFGIVVAATGTGGGVSISGKK951 pKa = 10.2GGNFTGKK958 pKa = 10.28AGGTHH963 pKa = 6.07TEE965 pKa = 3.71PNAYY969 pKa = 9.59VLAASGPISLTGTAGALSTTYY990 pKa = 11.37GDD992 pKa = 3.1VDD994 pKa = 4.42LNSTVGFVSTLPSGFGIASPVTSSSSNISITADD1027 pKa = 3.15SFKK1030 pKa = 10.41TDD1032 pKa = 3.22VVFGSGTFTSSAVQSSGTLSIAPRR1056 pKa = 11.84SPGKK1060 pKa = 10.38ALAVQTTPPSGTPLWINPTTMFGSSGLFKK1089 pKa = 10.46TGFSSFVFGSASTGTVTLDD1108 pKa = 2.83NYY1110 pKa = 10.67TFDD1113 pKa = 3.89NVTTVDD1119 pKa = 3.22TGANAILGTISIPNHH1134 pKa = 5.99ALTVNITGSGSVTDD1148 pKa = 3.57TGVIDD1153 pKa = 4.14VSSLALDD1160 pKa = 4.04AASSAVTLDD1169 pKa = 3.27NTGNAIGTLAADD1181 pKa = 3.9VASLSLDD1188 pKa = 3.22NATGLTVGSVAALNGVSATGGIDD1211 pKa = 3.01IEE1213 pKa = 4.57TATGSLTVTQNIATSDD1229 pKa = 3.5TGSAAIVLDD1238 pKa = 4.18AGNATAAGIATGGDD1252 pKa = 3.03IVLGGTPSPSISVGSGGKK1270 pKa = 9.6AVLYY1274 pKa = 8.73TGSVSGSTGIGALVGSGNSRR1294 pKa = 11.84YY1295 pKa = 9.97DD1296 pKa = 3.29SSKK1299 pKa = 7.27TTPGFTVALGSSGMFAVYY1317 pKa = 10.12RR1318 pKa = 11.84EE1319 pKa = 3.97QPILGYY1325 pKa = 8.24TAGSATITYY1334 pKa = 8.95GGSVPMSSLTPTYY1347 pKa = 10.53TSGLVNGDD1355 pKa = 3.45ALAQVVSSVPSASYY1369 pKa = 10.15TSSLSSGGYY1378 pKa = 8.96QNAGIYY1384 pKa = 9.92AVSASGGVTGFGYY1397 pKa = 10.58GFQYY1401 pKa = 10.62TGGTLTVNAKK1411 pKa = 10.32ALTITASAQSTTYY1424 pKa = 9.15GTALALGTSQFTPSGLVNGDD1444 pKa = 3.23SVAGVTLTQGGNATVPVTQAAGTYY1468 pKa = 8.31TGSTNGILASGATGTGLSNYY1488 pKa = 7.82TISYY1492 pKa = 9.98VPGTLTIDD1500 pKa = 3.82PKK1502 pKa = 11.25ALTVSGEE1509 pKa = 3.71AALNKK1514 pKa = 9.88VYY1516 pKa = 10.74DD1517 pKa = 4.1GSTTATLTGGTLPGLINGDD1536 pKa = 3.91SVTLTQAGTFASKK1549 pKa = 10.9NVGTNITVTASDD1561 pKa = 4.52SISGPSAGNYY1571 pKa = 8.76TLMQPTGLTANITQLASVTWIGGASGNWFDD1601 pKa = 3.78PANWAGGAVPDD1612 pKa = 4.64LSNVANVVIPQGVTVTFDD1630 pKa = 3.19SANLVAPAQGGPVNLASVGSAGNLTMAAGTLNVSSGVRR1668 pKa = 11.84LNTLAQTGGTVDD1680 pKa = 3.14SNGPVAVDD1688 pKa = 3.39VLTQSGGTLGGAGSVTASSLMQSGGSIVVGGNVTAGTLTQTAGSIDD1734 pKa = 3.28NGGNVGAVALAQSGGQIANRR1754 pKa = 11.84GGLDD1758 pKa = 3.26VTQSFSQSAPGSIAVGGSIAITQAAGNLSFVSLAGNDD1795 pKa = 2.91IDD1797 pKa = 5.75LGANSGAVNLGALDD1811 pKa = 3.77AAGSLTVNASGNISQQPGSVIRR1833 pKa = 11.84VATNVAMNSTAGAVTVSTAGNSFGSGLDD1861 pKa = 3.45SASPAQAAQSSVAADD1876 pKa = 3.43VEE1878 pKa = 4.73SGIEE1882 pKa = 3.95FTPEE1886 pKa = 3.22AGNLVEE1892 pKa = 4.78PALHH1896 pKa = 6.2TEE1898 pKa = 4.77PGQGMGTAGGSSEE1911 pKa = 3.95PAVAQEE1917 pKa = 4.22SGGAAVQPLAGIRR1930 pKa = 11.84LTVLDD1935 pKa = 3.96GGVRR1939 pKa = 11.84LPEE1942 pKa = 3.95LRR1944 pKa = 4.39
Molecular weight: 188.08 kDa
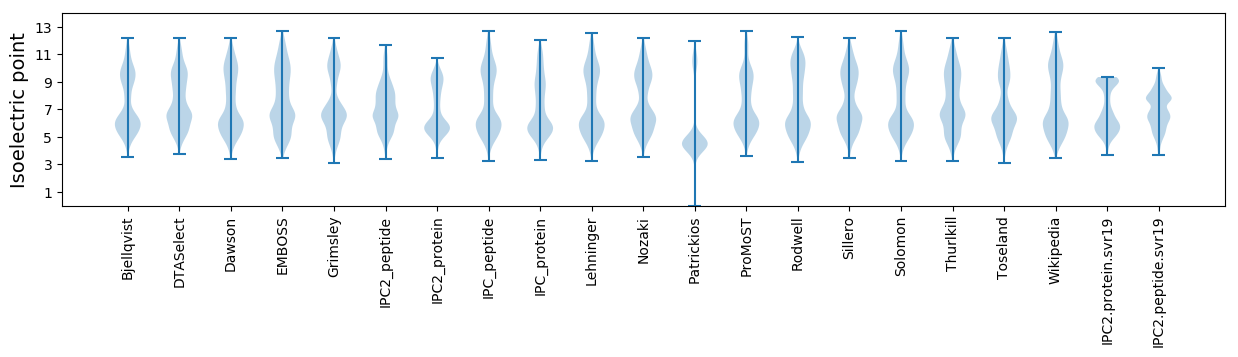
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1H2R9|A0A0A1H2R9_9BURK Bifunctional purine biosynthesis protein PurH OS=Burkholderiales bacterium GJ-E10 OX=1469502 GN=purH PE=3 SV=1
MM1 pKa = 8.06DD2 pKa = 5.77GIGWAWDD9 pKa = 3.6RR10 pKa = 11.84DD11 pKa = 4.2DD12 pKa = 4.75TDD14 pKa = 3.16TPYY17 pKa = 11.38AGLQQLAEE25 pKa = 4.25PPGEE29 pKa = 3.89YY30 pKa = 10.18SIFLATLHH38 pKa = 6.48PAGLSSSALIPINDD52 pKa = 3.54HH53 pKa = 5.92SAGVDD58 pKa = 3.79LSCHH62 pKa = 5.1IMARR66 pKa = 11.84SRR68 pKa = 11.84WGIVSADD75 pKa = 3.34PAGPLRR81 pKa = 11.84EE82 pKa = 3.89PAGRR86 pKa = 11.84VDD88 pKa = 3.66TGKK91 pKa = 10.11HH92 pKa = 4.95GMPIHH97 pKa = 6.16NAVIAATFTEE107 pKa = 4.36IADD110 pKa = 3.97LLEE113 pKa = 5.43IEE115 pKa = 4.37QANPFRR121 pKa = 11.84IGAYY125 pKa = 9.63RR126 pKa = 11.84KK127 pKa = 8.73AARR130 pKa = 11.84IARR133 pKa = 11.84ARR135 pKa = 11.84GLKK138 pKa = 9.96LNEE141 pKa = 3.79YY142 pKa = 9.9GVFRR146 pKa = 11.84GTEE149 pKa = 4.21RR150 pKa = 11.84IAGAAARR157 pKa = 11.84GGDD160 pKa = 3.41AVNWEE165 pKa = 4.55TFPHH169 pKa = 6.2EE170 pKa = 3.78ADD172 pKa = 2.91IGVRR176 pKa = 11.84GTGATMAEE184 pKa = 4.36AFAGAATALTAVVCDD199 pKa = 4.2PAAVGAAEE207 pKa = 4.3TTRR210 pKa = 11.84IEE212 pKa = 4.35CTAPDD217 pKa = 4.79DD218 pKa = 4.65EE219 pKa = 6.03LLLLDD224 pKa = 4.3WLNALIYY231 pKa = 10.68EE232 pKa = 4.33MATRR236 pKa = 11.84RR237 pKa = 11.84LLFSRR242 pKa = 11.84FAVRR246 pKa = 11.84ITGHH250 pKa = 6.46RR251 pKa = 11.84LRR253 pKa = 11.84ATAWGSTFPAACARR267 pKa = 11.84CIPASSAPTSSAKK280 pKa = 9.55RR281 pKa = 11.84RR282 pKa = 11.84RR283 pKa = 11.84WPISCLRR290 pKa = 11.84AFPPGSAAPAACVSMRR306 pKa = 11.84TEE308 pKa = 3.6WMRR311 pKa = 11.84CSKK314 pKa = 10.48AVRR317 pKa = 11.84AGPSSKK323 pKa = 10.72ASAVRR328 pKa = 11.84PTSRR332 pKa = 11.84ASRR335 pKa = 11.84STAASQAPIPARR347 pKa = 11.84FPSRR351 pKa = 11.84RR352 pKa = 11.84RR353 pKa = 11.84SASATRR359 pKa = 11.84WEE361 pKa = 4.29RR362 pKa = 11.84SVPATTISKK371 pKa = 10.14SRR373 pKa = 11.84SSKK376 pKa = 10.3PSSTPPRR383 pKa = 11.84RR384 pKa = 11.84APMAWRR390 pKa = 11.84RR391 pKa = 11.84TTWRR395 pKa = 3.45
MM1 pKa = 8.06DD2 pKa = 5.77GIGWAWDD9 pKa = 3.6RR10 pKa = 11.84DD11 pKa = 4.2DD12 pKa = 4.75TDD14 pKa = 3.16TPYY17 pKa = 11.38AGLQQLAEE25 pKa = 4.25PPGEE29 pKa = 3.89YY30 pKa = 10.18SIFLATLHH38 pKa = 6.48PAGLSSSALIPINDD52 pKa = 3.54HH53 pKa = 5.92SAGVDD58 pKa = 3.79LSCHH62 pKa = 5.1IMARR66 pKa = 11.84SRR68 pKa = 11.84WGIVSADD75 pKa = 3.34PAGPLRR81 pKa = 11.84EE82 pKa = 3.89PAGRR86 pKa = 11.84VDD88 pKa = 3.66TGKK91 pKa = 10.11HH92 pKa = 4.95GMPIHH97 pKa = 6.16NAVIAATFTEE107 pKa = 4.36IADD110 pKa = 3.97LLEE113 pKa = 5.43IEE115 pKa = 4.37QANPFRR121 pKa = 11.84IGAYY125 pKa = 9.63RR126 pKa = 11.84KK127 pKa = 8.73AARR130 pKa = 11.84IARR133 pKa = 11.84ARR135 pKa = 11.84GLKK138 pKa = 9.96LNEE141 pKa = 3.79YY142 pKa = 9.9GVFRR146 pKa = 11.84GTEE149 pKa = 4.21RR150 pKa = 11.84IAGAAARR157 pKa = 11.84GGDD160 pKa = 3.41AVNWEE165 pKa = 4.55TFPHH169 pKa = 6.2EE170 pKa = 3.78ADD172 pKa = 2.91IGVRR176 pKa = 11.84GTGATMAEE184 pKa = 4.36AFAGAATALTAVVCDD199 pKa = 4.2PAAVGAAEE207 pKa = 4.3TTRR210 pKa = 11.84IEE212 pKa = 4.35CTAPDD217 pKa = 4.79DD218 pKa = 4.65EE219 pKa = 6.03LLLLDD224 pKa = 4.3WLNALIYY231 pKa = 10.68EE232 pKa = 4.33MATRR236 pKa = 11.84RR237 pKa = 11.84LLFSRR242 pKa = 11.84FAVRR246 pKa = 11.84ITGHH250 pKa = 6.46RR251 pKa = 11.84LRR253 pKa = 11.84ATAWGSTFPAACARR267 pKa = 11.84CIPASSAPTSSAKK280 pKa = 9.55RR281 pKa = 11.84RR282 pKa = 11.84RR283 pKa = 11.84WPISCLRR290 pKa = 11.84AFPPGSAAPAACVSMRR306 pKa = 11.84TEE308 pKa = 3.6WMRR311 pKa = 11.84CSKK314 pKa = 10.48AVRR317 pKa = 11.84AGPSSKK323 pKa = 10.72ASAVRR328 pKa = 11.84PTSRR332 pKa = 11.84ASRR335 pKa = 11.84STAASQAPIPARR347 pKa = 11.84FPSRR351 pKa = 11.84RR352 pKa = 11.84RR353 pKa = 11.84SASATRR359 pKa = 11.84WEE361 pKa = 4.29RR362 pKa = 11.84SVPATTISKK371 pKa = 10.14SRR373 pKa = 11.84SSKK376 pKa = 10.3PSSTPPRR383 pKa = 11.84RR384 pKa = 11.84APMAWRR390 pKa = 11.84RR391 pKa = 11.84TTWRR395 pKa = 3.45
Molecular weight: 42.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
952706 |
40 |
7564 |
322.4 |
34.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.445 ± 0.075 | 0.966 ± 0.017 |
5.569 ± 0.033 | 5.411 ± 0.052 |
3.447 ± 0.033 | 8.412 ± 0.061 |
2.282 ± 0.024 | 4.711 ± 0.028 |
2.506 ± 0.037 | 10.096 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.227 ± 0.022 | 2.424 ± 0.036 |
5.457 ± 0.038 | 3.496 ± 0.028 |
8.263 ± 0.07 | 5.144 ± 0.064 |
5.023 ± 0.05 | 7.555 ± 0.044 |
1.374 ± 0.021 | 2.192 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
