
Microbacterium sp. RG1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
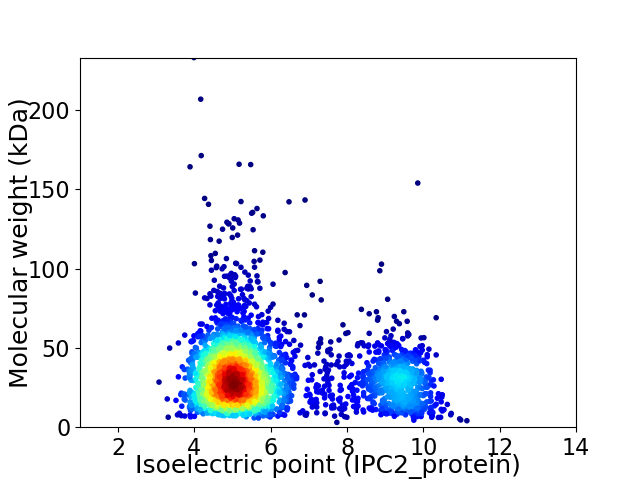
Virtual 2D-PAGE plot for 3028 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
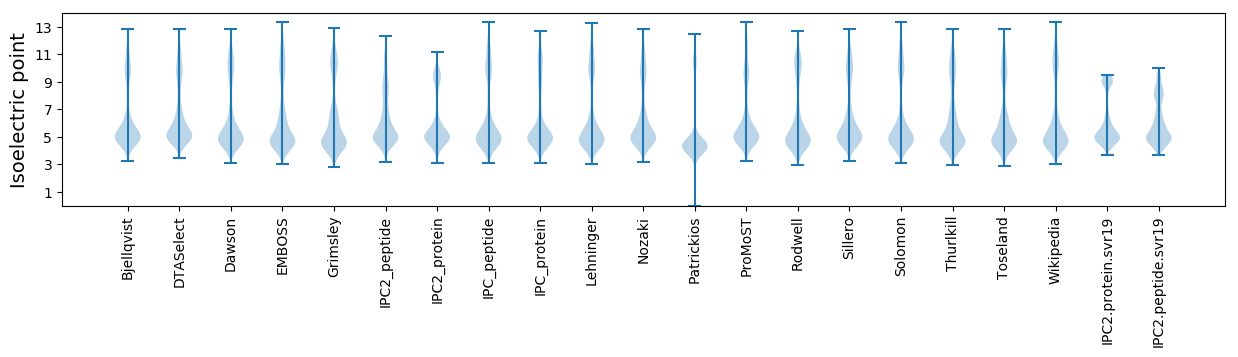
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8KMP2|A0A4P8KMP2_9MICO Phosphate transport system permease protein PstA OS=Microbacterium sp. RG1 OX=2489212 GN=pstA PE=3 SV=1
MM1 pKa = 7.79GIGIGAALAVTAGIAPAAAVDD22 pKa = 3.76PADD25 pKa = 3.64LEE27 pKa = 4.27EE28 pKa = 4.73RR29 pKa = 11.84VTVDD33 pKa = 3.08AVVGHH38 pKa = 6.58LQQLQDD44 pKa = 3.25IADD47 pKa = 3.81ANGGNRR53 pKa = 11.84AAGTSGYY60 pKa = 8.59EE61 pKa = 3.51ASAAYY66 pKa = 9.5IEE68 pKa = 4.35QALTAAGYY76 pKa = 8.21TPEE79 pKa = 4.16RR80 pKa = 11.84QYY82 pKa = 11.8FDD84 pKa = 5.51ADD86 pKa = 3.71LQTIDD91 pKa = 5.21AYY93 pKa = 9.93TLTVDD98 pKa = 4.81GYY100 pKa = 9.48TPADD104 pKa = 3.53PFGIPMEE111 pKa = 4.36FTPGTPAGGLTDD123 pKa = 4.91LPLIAPTAPLGCTGDD138 pKa = 3.65DD139 pKa = 3.43WAGTDD144 pKa = 3.34AAGAIALVSRR154 pKa = 11.84GSCSFAEE161 pKa = 4.44KK162 pKa = 10.24SAAAAAAGAEE172 pKa = 4.04AVIIYY177 pKa = 10.37NNEE180 pKa = 3.81AGPLQGTLGAQTPDD194 pKa = 3.19LAPTVGILQSEE205 pKa = 4.6GQDD208 pKa = 3.53LLAALADD215 pKa = 4.18GPVLADD221 pKa = 5.12LDD223 pKa = 4.25LQQHH227 pKa = 6.18IDD229 pKa = 3.4SVEE232 pKa = 4.03TFNIIADD239 pKa = 4.14TPTGRR244 pKa = 11.84DD245 pKa = 3.6DD246 pKa = 3.81NVVMLGAHH254 pKa = 6.98LDD256 pKa = 3.89SVEE259 pKa = 4.9DD260 pKa = 4.19GPGINDD266 pKa = 3.61NGSGSAAILEE276 pKa = 4.51VALQLAASGEE286 pKa = 4.3LEE288 pKa = 4.17NKK290 pKa = 9.21VRR292 pKa = 11.84FAWWGAEE299 pKa = 3.98EE300 pKa = 4.3IGLLGSAHH308 pKa = 6.15YY309 pKa = 10.78VDD311 pKa = 4.96EE312 pKa = 5.64LSDD315 pKa = 3.72ADD317 pKa = 3.75ADD319 pKa = 5.13AIATYY324 pKa = 11.15LNFDD328 pKa = 3.78MVASPNYY335 pKa = 9.76VIAVYY340 pKa = 10.45DD341 pKa = 4.08ADD343 pKa = 3.49QSTYY347 pKa = 9.03EE348 pKa = 4.34APVEE352 pKa = 4.16VPEE355 pKa = 4.7GSIATEE361 pKa = 3.9AAFTSYY367 pKa = 10.44FDD369 pKa = 6.05AIGQPWIDD377 pKa = 3.35TAFDD381 pKa = 3.23ARR383 pKa = 11.84SDD385 pKa = 3.73YY386 pKa = 11.37NAFILAGIPASGLFTGADD404 pKa = 3.36DD405 pKa = 4.08VKK407 pKa = 10.25TAEE410 pKa = 4.07QVALFGGTEE419 pKa = 4.43GIIMDD424 pKa = 4.89PNYY427 pKa = 8.05HH428 pKa = 6.05TPADD432 pKa = 3.87DD433 pKa = 3.39MSNINTEE440 pKa = 3.95ALGIMLGAMASVTAALANDD459 pKa = 4.1TSAVNGVAPIVDD471 pKa = 4.3PVAPAPEE478 pKa = 4.11VTEE481 pKa = 4.54EE482 pKa = 4.3GDD484 pKa = 3.59HH485 pKa = 6.81LLAASGSEE493 pKa = 4.08ISGVWLGGGVLALLAGAAVLAAAARR518 pKa = 11.84RR519 pKa = 11.84RR520 pKa = 11.84RR521 pKa = 11.84TVV523 pKa = 2.49
MM1 pKa = 7.79GIGIGAALAVTAGIAPAAAVDD22 pKa = 3.76PADD25 pKa = 3.64LEE27 pKa = 4.27EE28 pKa = 4.73RR29 pKa = 11.84VTVDD33 pKa = 3.08AVVGHH38 pKa = 6.58LQQLQDD44 pKa = 3.25IADD47 pKa = 3.81ANGGNRR53 pKa = 11.84AAGTSGYY60 pKa = 8.59EE61 pKa = 3.51ASAAYY66 pKa = 9.5IEE68 pKa = 4.35QALTAAGYY76 pKa = 8.21TPEE79 pKa = 4.16RR80 pKa = 11.84QYY82 pKa = 11.8FDD84 pKa = 5.51ADD86 pKa = 3.71LQTIDD91 pKa = 5.21AYY93 pKa = 9.93TLTVDD98 pKa = 4.81GYY100 pKa = 9.48TPADD104 pKa = 3.53PFGIPMEE111 pKa = 4.36FTPGTPAGGLTDD123 pKa = 4.91LPLIAPTAPLGCTGDD138 pKa = 3.65DD139 pKa = 3.43WAGTDD144 pKa = 3.34AAGAIALVSRR154 pKa = 11.84GSCSFAEE161 pKa = 4.44KK162 pKa = 10.24SAAAAAAGAEE172 pKa = 4.04AVIIYY177 pKa = 10.37NNEE180 pKa = 3.81AGPLQGTLGAQTPDD194 pKa = 3.19LAPTVGILQSEE205 pKa = 4.6GQDD208 pKa = 3.53LLAALADD215 pKa = 4.18GPVLADD221 pKa = 5.12LDD223 pKa = 4.25LQQHH227 pKa = 6.18IDD229 pKa = 3.4SVEE232 pKa = 4.03TFNIIADD239 pKa = 4.14TPTGRR244 pKa = 11.84DD245 pKa = 3.6DD246 pKa = 3.81NVVMLGAHH254 pKa = 6.98LDD256 pKa = 3.89SVEE259 pKa = 4.9DD260 pKa = 4.19GPGINDD266 pKa = 3.61NGSGSAAILEE276 pKa = 4.51VALQLAASGEE286 pKa = 4.3LEE288 pKa = 4.17NKK290 pKa = 9.21VRR292 pKa = 11.84FAWWGAEE299 pKa = 3.98EE300 pKa = 4.3IGLLGSAHH308 pKa = 6.15YY309 pKa = 10.78VDD311 pKa = 4.96EE312 pKa = 5.64LSDD315 pKa = 3.72ADD317 pKa = 3.75ADD319 pKa = 5.13AIATYY324 pKa = 11.15LNFDD328 pKa = 3.78MVASPNYY335 pKa = 9.76VIAVYY340 pKa = 10.45DD341 pKa = 4.08ADD343 pKa = 3.49QSTYY347 pKa = 9.03EE348 pKa = 4.34APVEE352 pKa = 4.16VPEE355 pKa = 4.7GSIATEE361 pKa = 3.9AAFTSYY367 pKa = 10.44FDD369 pKa = 6.05AIGQPWIDD377 pKa = 3.35TAFDD381 pKa = 3.23ARR383 pKa = 11.84SDD385 pKa = 3.73YY386 pKa = 11.37NAFILAGIPASGLFTGADD404 pKa = 3.36DD405 pKa = 4.08VKK407 pKa = 10.25TAEE410 pKa = 4.07QVALFGGTEE419 pKa = 4.43GIIMDD424 pKa = 4.89PNYY427 pKa = 8.05HH428 pKa = 6.05TPADD432 pKa = 3.87DD433 pKa = 3.39MSNINTEE440 pKa = 3.95ALGIMLGAMASVTAALANDD459 pKa = 4.1TSAVNGVAPIVDD471 pKa = 4.3PVAPAPEE478 pKa = 4.11VTEE481 pKa = 4.54EE482 pKa = 4.3GDD484 pKa = 3.59HH485 pKa = 6.81LLAASGSEE493 pKa = 4.08ISGVWLGGGVLALLAGAAVLAAAARR518 pKa = 11.84RR519 pKa = 11.84RR520 pKa = 11.84RR521 pKa = 11.84TVV523 pKa = 2.49
Molecular weight: 53.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1EQD3|A0A4V1EQD3_9MICO Nucleotidyltransferase family protein OS=Microbacterium sp. RG1 OX=2489212 GN=EHF32_11790 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
979390 |
29 |
2287 |
323.4 |
34.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.147 ± 0.064 | 0.471 ± 0.01 |
6.212 ± 0.045 | 5.493 ± 0.045 |
3.094 ± 0.028 | 8.832 ± 0.041 |
1.893 ± 0.022 | 4.474 ± 0.03 |
1.726 ± 0.03 | 10.173 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.751 ± 0.019 | 1.846 ± 0.025 |
5.349 ± 0.03 | 2.746 ± 0.023 |
7.556 ± 0.058 | 5.614 ± 0.028 |
6.012 ± 0.043 | 9.124 ± 0.042 |
1.531 ± 0.019 | 1.956 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
