
Red clover necrotic mosaic virus (RCNMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Regressovirinae; Dianthovirus
Average proteome isoelectric point is 7.98
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
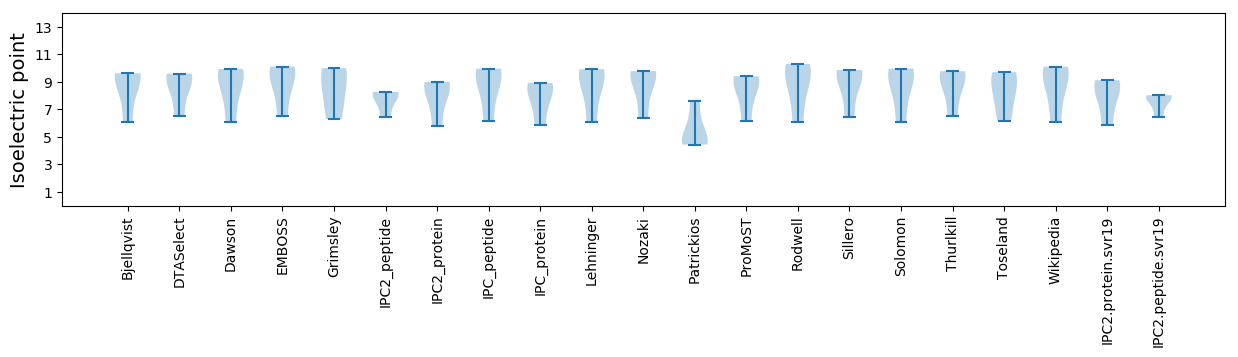
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P22956-2|RDRP-2_RCNMV Isoform of P22956 Isoform protein p27 of RNA-directed RNA polymerase OS=Red clover necrotic mosaic virus OX=12267 GN=ORF1 PE=1 SV=2
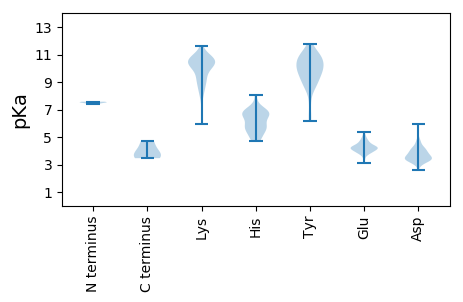
MM1 pKa = 7.52GFINLSLFDD10 pKa = 3.87VDD12 pKa = 5.98KK13 pKa = 11.65LMVWVSKK20 pKa = 9.71FNPGKK25 pKa = 10.11ILSAICNLGIDD36 pKa = 3.47CWNRR40 pKa = 11.84FRR42 pKa = 11.84KK43 pKa = 8.86WFFGLNFDD51 pKa = 3.42AHH53 pKa = 5.56MWAVDD58 pKa = 3.32AFIPLMPHH66 pKa = 4.81YY67 pKa = 8.64TEE69 pKa = 3.78QMEE72 pKa = 4.26RR73 pKa = 11.84VVDD76 pKa = 4.17DD77 pKa = 4.66FCSEE81 pKa = 4.14TPEE84 pKa = 4.87SKK86 pKa = 11.11LEE88 pKa = 4.29DD89 pKa = 4.15CLEE92 pKa = 4.65LDD94 pKa = 3.49TSVNEE99 pKa = 4.22FFDD102 pKa = 3.83EE103 pKa = 4.11EE104 pKa = 5.0VYY106 pKa = 10.98KK107 pKa = 10.62KK108 pKa = 10.67DD109 pKa = 3.34EE110 pKa = 4.42EE111 pKa = 4.77GVMKK115 pKa = 10.25LQRR118 pKa = 11.84SAARR122 pKa = 11.84KK123 pKa = 7.86HH124 pKa = 5.69IKK126 pKa = 9.67RR127 pKa = 11.84VRR129 pKa = 11.84PGMMQAAIKK138 pKa = 10.36AVEE141 pKa = 3.71TRR143 pKa = 11.84IRR145 pKa = 11.84NRR147 pKa = 11.84HH148 pKa = 5.4TIFGDD153 pKa = 3.44DD154 pKa = 3.3MGKK157 pKa = 9.81VDD159 pKa = 4.15EE160 pKa = 4.57AAVRR164 pKa = 11.84ATASDD169 pKa = 2.96ICGEE173 pKa = 4.01FKK175 pKa = 10.74INEE178 pKa = 4.14HH179 pKa = 5.86HH180 pKa = 6.76TNALVYY186 pKa = 9.94AAAYY190 pKa = 10.28LAMTPDD196 pKa = 3.28QRR198 pKa = 11.84SIDD201 pKa = 4.17SVKK204 pKa = 10.37LAYY207 pKa = 9.99NPKK210 pKa = 8.46SQARR214 pKa = 11.84RR215 pKa = 11.84TLVSAIRR222 pKa = 11.84EE223 pKa = 4.18NKK225 pKa = 9.26AVAGFKK231 pKa = 10.61SLEE234 pKa = 4.18DD235 pKa = 3.79FF236 pKa = 4.71
MM1 pKa = 7.52GFINLSLFDD10 pKa = 3.87VDD12 pKa = 5.98KK13 pKa = 11.65LMVWVSKK20 pKa = 9.71FNPGKK25 pKa = 10.11ILSAICNLGIDD36 pKa = 3.47CWNRR40 pKa = 11.84FRR42 pKa = 11.84KK43 pKa = 8.86WFFGLNFDD51 pKa = 3.42AHH53 pKa = 5.56MWAVDD58 pKa = 3.32AFIPLMPHH66 pKa = 4.81YY67 pKa = 8.64TEE69 pKa = 3.78QMEE72 pKa = 4.26RR73 pKa = 11.84VVDD76 pKa = 4.17DD77 pKa = 4.66FCSEE81 pKa = 4.14TPEE84 pKa = 4.87SKK86 pKa = 11.11LEE88 pKa = 4.29DD89 pKa = 4.15CLEE92 pKa = 4.65LDD94 pKa = 3.49TSVNEE99 pKa = 4.22FFDD102 pKa = 3.83EE103 pKa = 4.11EE104 pKa = 5.0VYY106 pKa = 10.98KK107 pKa = 10.62KK108 pKa = 10.67DD109 pKa = 3.34EE110 pKa = 4.42EE111 pKa = 4.77GVMKK115 pKa = 10.25LQRR118 pKa = 11.84SAARR122 pKa = 11.84KK123 pKa = 7.86HH124 pKa = 5.69IKK126 pKa = 9.67RR127 pKa = 11.84VRR129 pKa = 11.84PGMMQAAIKK138 pKa = 10.36AVEE141 pKa = 3.71TRR143 pKa = 11.84IRR145 pKa = 11.84NRR147 pKa = 11.84HH148 pKa = 5.4TIFGDD153 pKa = 3.44DD154 pKa = 3.3MGKK157 pKa = 9.81VDD159 pKa = 4.15EE160 pKa = 4.57AAVRR164 pKa = 11.84ATASDD169 pKa = 2.96ICGEE173 pKa = 4.01FKK175 pKa = 10.74INEE178 pKa = 4.14HH179 pKa = 5.86HH180 pKa = 6.76TNALVYY186 pKa = 9.94AAAYY190 pKa = 10.28LAMTPDD196 pKa = 3.28QRR198 pKa = 11.84SIDD201 pKa = 4.17SVKK204 pKa = 10.37LAYY207 pKa = 9.99NPKK210 pKa = 8.46SQARR214 pKa = 11.84RR215 pKa = 11.84TLVSAIRR222 pKa = 11.84EE223 pKa = 4.18NKK225 pKa = 9.26AVAGFKK231 pKa = 10.61SLEE234 pKa = 4.18DD235 pKa = 3.79FF236 pKa = 4.71
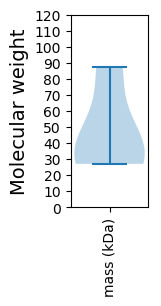
Molecular weight: 27.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P22955|CAPSD_RCNMV Capsid protein OS=Red clover necrotic mosaic virus OX=12267 GN=ORF2 PE=1 SV=1
MM1 pKa = 7.4AVHH4 pKa = 6.32VEE6 pKa = 4.09NLSDD10 pKa = 4.26LAKK13 pKa = 10.18TNDD16 pKa = 3.45GVAVSLNRR24 pKa = 11.84YY25 pKa = 6.56TDD27 pKa = 3.42WKK29 pKa = 9.81CRR31 pKa = 11.84SGVSEE36 pKa = 4.77APLIPASMMSKK47 pKa = 8.26ITDD50 pKa = 3.71YY51 pKa = 11.79AKK53 pKa = 8.35TTAKK57 pKa = 10.38GNSVALNYY65 pKa = 8.76THH67 pKa = 6.73VVLSLAPTIGVAIPGHH83 pKa = 4.75VTVEE87 pKa = 4.4LINPNVEE94 pKa = 3.93GPFQVMSGQTLSWSPGAGKK113 pKa = 8.96PCLMIFSVHH122 pKa = 5.83HH123 pKa = 5.56QLNSDD128 pKa = 3.73HH129 pKa = 6.98EE130 pKa = 4.32PFRR133 pKa = 11.84VRR135 pKa = 11.84ITNTGIPTKK144 pKa = 10.28KK145 pKa = 10.28SYY147 pKa = 10.75ARR149 pKa = 11.84CHH151 pKa = 6.39AYY153 pKa = 9.08WGFDD157 pKa = 2.58VGTRR161 pKa = 11.84HH162 pKa = 6.72RR163 pKa = 11.84YY164 pKa = 8.8YY165 pKa = 10.47KK166 pKa = 10.32SEE168 pKa = 3.93PARR171 pKa = 11.84LIEE174 pKa = 5.18LEE176 pKa = 3.93VGYY179 pKa = 10.52QRR181 pKa = 11.84TLLSSIKK188 pKa = 10.41AVEE191 pKa = 4.51AYY193 pKa = 10.42VQFTFDD199 pKa = 3.41TSRR202 pKa = 11.84MEE204 pKa = 5.38KK205 pKa = 10.36NPQLCTKK212 pKa = 10.03SNVNIIPPKK221 pKa = 10.59AEE223 pKa = 3.82TGSIRR228 pKa = 11.84GIAPPLSVVPNQGRR242 pKa = 11.84EE243 pKa = 4.25SKK245 pKa = 10.62VLKK248 pKa = 10.48QKK250 pKa = 10.92GGTGSKK256 pKa = 5.98TTKK259 pKa = 10.37LPSLEE264 pKa = 4.29PSSGSSSGLSMSRR277 pKa = 11.84RR278 pKa = 11.84SHH280 pKa = 6.69RR281 pKa = 11.84NVLNSSIPIKK291 pKa = 10.58RR292 pKa = 11.84NQDD295 pKa = 3.45GNWLGDD301 pKa = 3.59HH302 pKa = 6.93LSDD305 pKa = 4.58KK306 pKa = 11.22GRR308 pKa = 11.84VTDD311 pKa = 4.64PNPEE315 pKa = 4.01RR316 pKa = 11.84LL317 pKa = 3.58
MM1 pKa = 7.4AVHH4 pKa = 6.32VEE6 pKa = 4.09NLSDD10 pKa = 4.26LAKK13 pKa = 10.18TNDD16 pKa = 3.45GVAVSLNRR24 pKa = 11.84YY25 pKa = 6.56TDD27 pKa = 3.42WKK29 pKa = 9.81CRR31 pKa = 11.84SGVSEE36 pKa = 4.77APLIPASMMSKK47 pKa = 8.26ITDD50 pKa = 3.71YY51 pKa = 11.79AKK53 pKa = 8.35TTAKK57 pKa = 10.38GNSVALNYY65 pKa = 8.76THH67 pKa = 6.73VVLSLAPTIGVAIPGHH83 pKa = 4.75VTVEE87 pKa = 4.4LINPNVEE94 pKa = 3.93GPFQVMSGQTLSWSPGAGKK113 pKa = 8.96PCLMIFSVHH122 pKa = 5.83HH123 pKa = 5.56QLNSDD128 pKa = 3.73HH129 pKa = 6.98EE130 pKa = 4.32PFRR133 pKa = 11.84VRR135 pKa = 11.84ITNTGIPTKK144 pKa = 10.28KK145 pKa = 10.28SYY147 pKa = 10.75ARR149 pKa = 11.84CHH151 pKa = 6.39AYY153 pKa = 9.08WGFDD157 pKa = 2.58VGTRR161 pKa = 11.84HH162 pKa = 6.72RR163 pKa = 11.84YY164 pKa = 8.8YY165 pKa = 10.47KK166 pKa = 10.32SEE168 pKa = 3.93PARR171 pKa = 11.84LIEE174 pKa = 5.18LEE176 pKa = 3.93VGYY179 pKa = 10.52QRR181 pKa = 11.84TLLSSIKK188 pKa = 10.41AVEE191 pKa = 4.51AYY193 pKa = 10.42VQFTFDD199 pKa = 3.41TSRR202 pKa = 11.84MEE204 pKa = 5.38KK205 pKa = 10.36NPQLCTKK212 pKa = 10.03SNVNIIPPKK221 pKa = 10.59AEE223 pKa = 3.82TGSIRR228 pKa = 11.84GIAPPLSVVPNQGRR242 pKa = 11.84EE243 pKa = 4.25SKK245 pKa = 10.62VLKK248 pKa = 10.48QKK250 pKa = 10.92GGTGSKK256 pKa = 5.98TTKK259 pKa = 10.37LPSLEE264 pKa = 4.29PSSGSSSGLSMSRR277 pKa = 11.84RR278 pKa = 11.84SHH280 pKa = 6.69RR281 pKa = 11.84NVLNSSIPIKK291 pKa = 10.58RR292 pKa = 11.84NQDD295 pKa = 3.45GNWLGDD301 pKa = 3.59HH302 pKa = 6.93LSDD305 pKa = 4.58KK306 pKa = 11.22GRR308 pKa = 11.84VTDD311 pKa = 4.64PNPEE315 pKa = 4.01RR316 pKa = 11.84LL317 pKa = 3.58
Molecular weight: 34.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1659 |
236 |
767 |
414.8 |
46.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.113 ± 0.673 | 2.049 ± 0.324 |
5.425 ± 0.68 | 5.184 ± 0.885 |
4.34 ± 0.669 | 6.088 ± 0.606 |
2.11 ± 0.354 | 5.184 ± 0.135 |
6.751 ± 0.612 | 7.957 ± 0.503 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.773 ± 0.352 | 4.702 ± 0.26 |
4.762 ± 0.772 | 3.737 ± 0.443 |
6.269 ± 0.46 | 8.258 ± 1.349 |
5.546 ± 1.203 | 7.715 ± 0.475 |
1.386 ± 0.221 | 2.652 ± 0.13 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
