
Pseudomonas sp. StFLB209
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; unclassified Pseudomonas
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
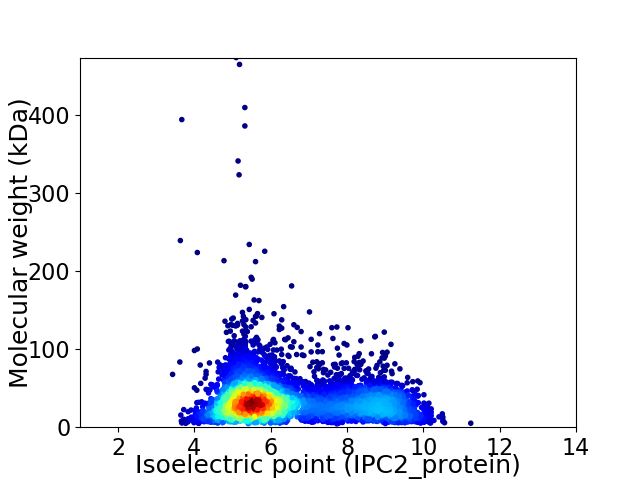
Virtual 2D-PAGE plot for 5594 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A077LJQ1|A0A077LJQ1_9PSED Imidazoleglycerol-phosphate dehydratase OS=Pseudomonas sp. StFLB209 OX=1028989 GN=hisB PE=3 SV=1
MM1 pKa = 7.39AVINGTSGADD11 pKa = 3.47TLTGTSDD18 pKa = 3.2SDD20 pKa = 4.23QINGLAGNDD29 pKa = 4.0VIHH32 pKa = 6.92SSAGADD38 pKa = 3.44KK39 pKa = 10.71IVGGVGTDD47 pKa = 3.07TVDD50 pKa = 3.33YY51 pKa = 10.2SKK53 pKa = 10.9SAEE56 pKa = 4.45GISIDD61 pKa = 3.35VRR63 pKa = 11.84AGVGLAGRR71 pKa = 11.84GGDD74 pKa = 3.51AEE76 pKa = 4.53GDD78 pKa = 3.27ILEE81 pKa = 4.47TVEE84 pKa = 4.4KK85 pKa = 10.96VVGSVFNDD93 pKa = 3.42TLAIGAIATMVTLEE107 pKa = 4.6GGAGDD112 pKa = 3.61DD113 pKa = 4.09TYY115 pKa = 11.05IIGHH119 pKa = 7.58DD120 pKa = 3.58GTPIIVEE127 pKa = 4.04QANGGNDD134 pKa = 3.49EE135 pKa = 4.11VRR137 pKa = 11.84VTVLSKK143 pKa = 11.07QNTRR147 pKa = 11.84LADD150 pKa = 3.96NIEE153 pKa = 3.88RR154 pKa = 11.84LTYY157 pKa = 10.09VGNGAFTGYY166 pKa = 10.86GNALDD171 pKa = 4.7NIITGGSGGDD181 pKa = 3.44KK182 pKa = 10.64LYY184 pKa = 11.27GGAGADD190 pKa = 3.55QFIGGAGNDD199 pKa = 3.53TVGYY203 pKa = 10.07DD204 pKa = 3.69DD205 pKa = 3.9STTGVSINLKK215 pKa = 8.88TGEE218 pKa = 4.33KK219 pKa = 10.15SGIAAGDD226 pKa = 3.76TYY228 pKa = 11.54TDD230 pKa = 3.75IEE232 pKa = 4.81VIWGSNANDD241 pKa = 3.68TFVGDD246 pKa = 3.9GSGMDD251 pKa = 3.82FNGLGGIDD259 pKa = 3.39TVDD262 pKa = 3.49YY263 pKa = 10.6SGSASAVDD271 pKa = 3.57VSVSVGNVRR280 pKa = 11.84AGIGGDD286 pKa = 3.56AEE288 pKa = 4.3GTVLNNIEE296 pKa = 4.76RR297 pKa = 11.84IVGSAFNDD305 pKa = 3.44TLAIGTISTMVTLEE319 pKa = 4.21GGAGDD324 pKa = 3.61DD325 pKa = 4.06TYY327 pKa = 11.05IIGHH331 pKa = 6.67NGTPIVVEE339 pKa = 3.96QANGGNDD346 pKa = 3.49EE347 pKa = 4.23VRR349 pKa = 11.84VTVLNQQGTRR359 pKa = 11.84LADD362 pKa = 3.86NIEE365 pKa = 3.88RR366 pKa = 11.84LTYY369 pKa = 10.09VGNGAFTGYY378 pKa = 10.86GNALDD383 pKa = 4.7NIITGGSGNNTLYY396 pKa = 11.04GGAGDD401 pKa = 4.06DD402 pKa = 4.56QIIGGAGSDD411 pKa = 3.55TLNGGEE417 pKa = 5.21GADD420 pKa = 3.71QLIGGAGYY428 pKa = 7.99DD429 pKa = 3.45TAGYY433 pKa = 10.13ADD435 pKa = 4.88SISAGVTVNLKK446 pKa = 9.43TGIHH450 pKa = 6.08SGFAAGDD457 pKa = 3.66TYY459 pKa = 11.83VDD461 pKa = 3.51IEE463 pKa = 5.03AISGSNFNDD472 pKa = 2.84VFYY475 pKa = 10.96ADD477 pKa = 4.01SSSTGWNGVVGQDD490 pKa = 2.99MVTYY494 pKa = 8.78EE495 pKa = 4.17LSEE498 pKa = 4.14TAVTVDD504 pKa = 5.12LFTGTNAGDD513 pKa = 3.91ADD515 pKa = 4.07GDD517 pKa = 4.1TYY519 pKa = 11.85ASIEE523 pKa = 4.21IIQGSTFGDD532 pKa = 3.67TLSGSRR538 pKa = 11.84LADD541 pKa = 3.43TFIGGAGADD550 pKa = 3.9TIDD553 pKa = 3.43GRR555 pKa = 11.84EE556 pKa = 3.99GHH558 pKa = 6.89DD559 pKa = 3.7SVWYY563 pKa = 7.34ITSAAGVTINLQTQSNQGGDD583 pKa = 3.33AEE585 pKa = 5.09GDD587 pKa = 3.52VLSNIEE593 pKa = 4.28RR594 pKa = 11.84LVGSHH599 pKa = 7.0FNDD602 pKa = 4.12ALTGDD607 pKa = 3.53AGVNYY612 pKa = 10.59LEE614 pKa = 5.48GGLGDD619 pKa = 5.39DD620 pKa = 4.46IIEE623 pKa = 4.48GGEE626 pKa = 3.94GGDD629 pKa = 3.76YY630 pKa = 10.77IFGGLASQIGSFTLVGQTNGPQADD654 pKa = 3.67TLYY657 pKa = 11.01GGNGNDD663 pKa = 4.37TIVTAANDD671 pKa = 3.68LGSRR675 pKa = 11.84VYY677 pKa = 11.01GEE679 pKa = 4.51AGGDD683 pKa = 3.75VITVVHH689 pKa = 6.26GMADD693 pKa = 3.36GGDD696 pKa = 3.93GNDD699 pKa = 3.2VLTGTGLDD707 pKa = 3.65FSLFGGSGSDD717 pKa = 3.15QLVLHH722 pKa = 6.39SSGWANGGEE731 pKa = 4.3GSDD734 pKa = 3.69TYY736 pKa = 9.62TVRR739 pKa = 11.84TTKK742 pKa = 10.76LVMIQDD748 pKa = 4.51DD749 pKa = 4.15GASYY753 pKa = 11.27GDD755 pKa = 4.24KK756 pKa = 10.82LVLSQITSSEE766 pKa = 4.1LLFDD770 pKa = 4.52QIGNDD775 pKa = 3.87LYY777 pKa = 11.08LHH779 pKa = 7.07RR780 pKa = 11.84YY781 pKa = 8.48SLAAGQTPQEE791 pKa = 4.18GVLLKK796 pKa = 10.69DD797 pKa = 3.39WFAGYY802 pKa = 8.27DD803 pKa = 3.81TIEE806 pKa = 4.34HH807 pKa = 5.67VQTADD812 pKa = 3.16MQLISLSGLSPTTSSLAA829 pKa = 3.44
MM1 pKa = 7.39AVINGTSGADD11 pKa = 3.47TLTGTSDD18 pKa = 3.2SDD20 pKa = 4.23QINGLAGNDD29 pKa = 4.0VIHH32 pKa = 6.92SSAGADD38 pKa = 3.44KK39 pKa = 10.71IVGGVGTDD47 pKa = 3.07TVDD50 pKa = 3.33YY51 pKa = 10.2SKK53 pKa = 10.9SAEE56 pKa = 4.45GISIDD61 pKa = 3.35VRR63 pKa = 11.84AGVGLAGRR71 pKa = 11.84GGDD74 pKa = 3.51AEE76 pKa = 4.53GDD78 pKa = 3.27ILEE81 pKa = 4.47TVEE84 pKa = 4.4KK85 pKa = 10.96VVGSVFNDD93 pKa = 3.42TLAIGAIATMVTLEE107 pKa = 4.6GGAGDD112 pKa = 3.61DD113 pKa = 4.09TYY115 pKa = 11.05IIGHH119 pKa = 7.58DD120 pKa = 3.58GTPIIVEE127 pKa = 4.04QANGGNDD134 pKa = 3.49EE135 pKa = 4.11VRR137 pKa = 11.84VTVLSKK143 pKa = 11.07QNTRR147 pKa = 11.84LADD150 pKa = 3.96NIEE153 pKa = 3.88RR154 pKa = 11.84LTYY157 pKa = 10.09VGNGAFTGYY166 pKa = 10.86GNALDD171 pKa = 4.7NIITGGSGGDD181 pKa = 3.44KK182 pKa = 10.64LYY184 pKa = 11.27GGAGADD190 pKa = 3.55QFIGGAGNDD199 pKa = 3.53TVGYY203 pKa = 10.07DD204 pKa = 3.69DD205 pKa = 3.9STTGVSINLKK215 pKa = 8.88TGEE218 pKa = 4.33KK219 pKa = 10.15SGIAAGDD226 pKa = 3.76TYY228 pKa = 11.54TDD230 pKa = 3.75IEE232 pKa = 4.81VIWGSNANDD241 pKa = 3.68TFVGDD246 pKa = 3.9GSGMDD251 pKa = 3.82FNGLGGIDD259 pKa = 3.39TVDD262 pKa = 3.49YY263 pKa = 10.6SGSASAVDD271 pKa = 3.57VSVSVGNVRR280 pKa = 11.84AGIGGDD286 pKa = 3.56AEE288 pKa = 4.3GTVLNNIEE296 pKa = 4.76RR297 pKa = 11.84IVGSAFNDD305 pKa = 3.44TLAIGTISTMVTLEE319 pKa = 4.21GGAGDD324 pKa = 3.61DD325 pKa = 4.06TYY327 pKa = 11.05IIGHH331 pKa = 6.67NGTPIVVEE339 pKa = 3.96QANGGNDD346 pKa = 3.49EE347 pKa = 4.23VRR349 pKa = 11.84VTVLNQQGTRR359 pKa = 11.84LADD362 pKa = 3.86NIEE365 pKa = 3.88RR366 pKa = 11.84LTYY369 pKa = 10.09VGNGAFTGYY378 pKa = 10.86GNALDD383 pKa = 4.7NIITGGSGNNTLYY396 pKa = 11.04GGAGDD401 pKa = 4.06DD402 pKa = 4.56QIIGGAGSDD411 pKa = 3.55TLNGGEE417 pKa = 5.21GADD420 pKa = 3.71QLIGGAGYY428 pKa = 7.99DD429 pKa = 3.45TAGYY433 pKa = 10.13ADD435 pKa = 4.88SISAGVTVNLKK446 pKa = 9.43TGIHH450 pKa = 6.08SGFAAGDD457 pKa = 3.66TYY459 pKa = 11.83VDD461 pKa = 3.51IEE463 pKa = 5.03AISGSNFNDD472 pKa = 2.84VFYY475 pKa = 10.96ADD477 pKa = 4.01SSSTGWNGVVGQDD490 pKa = 2.99MVTYY494 pKa = 8.78EE495 pKa = 4.17LSEE498 pKa = 4.14TAVTVDD504 pKa = 5.12LFTGTNAGDD513 pKa = 3.91ADD515 pKa = 4.07GDD517 pKa = 4.1TYY519 pKa = 11.85ASIEE523 pKa = 4.21IIQGSTFGDD532 pKa = 3.67TLSGSRR538 pKa = 11.84LADD541 pKa = 3.43TFIGGAGADD550 pKa = 3.9TIDD553 pKa = 3.43GRR555 pKa = 11.84EE556 pKa = 3.99GHH558 pKa = 6.89DD559 pKa = 3.7SVWYY563 pKa = 7.34ITSAAGVTINLQTQSNQGGDD583 pKa = 3.33AEE585 pKa = 5.09GDD587 pKa = 3.52VLSNIEE593 pKa = 4.28RR594 pKa = 11.84LVGSHH599 pKa = 7.0FNDD602 pKa = 4.12ALTGDD607 pKa = 3.53AGVNYY612 pKa = 10.59LEE614 pKa = 5.48GGLGDD619 pKa = 5.39DD620 pKa = 4.46IIEE623 pKa = 4.48GGEE626 pKa = 3.94GGDD629 pKa = 3.76YY630 pKa = 10.77IFGGLASQIGSFTLVGQTNGPQADD654 pKa = 3.67TLYY657 pKa = 11.01GGNGNDD663 pKa = 4.37TIVTAANDD671 pKa = 3.68LGSRR675 pKa = 11.84VYY677 pKa = 11.01GEE679 pKa = 4.51AGGDD683 pKa = 3.75VITVVHH689 pKa = 6.26GMADD693 pKa = 3.36GGDD696 pKa = 3.93GNDD699 pKa = 3.2VLTGTGLDD707 pKa = 3.65FSLFGGSGSDD717 pKa = 3.15QLVLHH722 pKa = 6.39SSGWANGGEE731 pKa = 4.3GSDD734 pKa = 3.69TYY736 pKa = 9.62TVRR739 pKa = 11.84TTKK742 pKa = 10.76LVMIQDD748 pKa = 4.51DD749 pKa = 4.15GASYY753 pKa = 11.27GDD755 pKa = 4.24KK756 pKa = 10.82LVLSQITSSEE766 pKa = 4.1LLFDD770 pKa = 4.52QIGNDD775 pKa = 3.87LYY777 pKa = 11.08LHH779 pKa = 7.07RR780 pKa = 11.84YY781 pKa = 8.48SLAAGQTPQEE791 pKa = 4.18GVLLKK796 pKa = 10.69DD797 pKa = 3.39WFAGYY802 pKa = 8.27DD803 pKa = 3.81TIEE806 pKa = 4.34HH807 pKa = 5.67VQTADD812 pKa = 3.16MQLISLSGLSPTTSSLAA829 pKa = 3.44
Molecular weight: 83.63 kDa
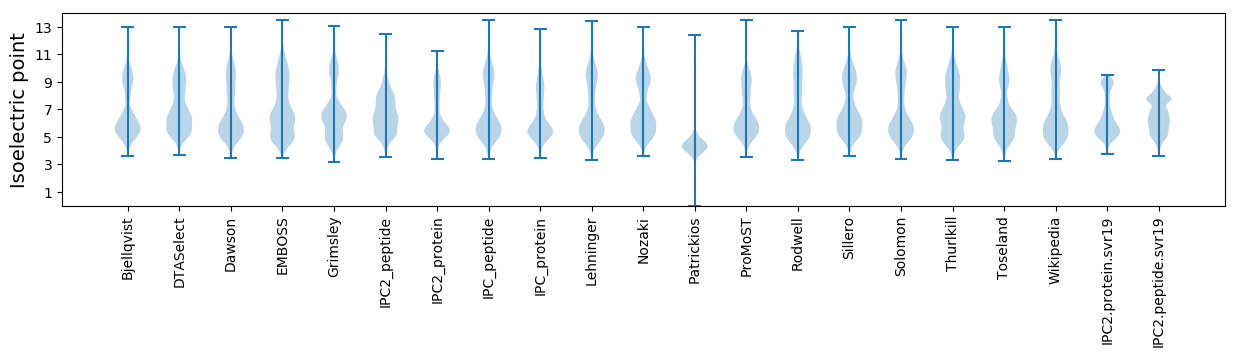
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077LKU1|A0A077LKU1_9PSED Uncharacterized protein OS=Pseudomonas sp. StFLB209 OX=1028989 GN=PSCI_4059 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTIKK11 pKa = 10.52RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.98GRR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84LAII44 pKa = 4.0
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTIKK11 pKa = 10.52RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.98GRR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84LAII44 pKa = 4.0
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1859865 |
39 |
4311 |
332.5 |
36.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.093 ± 0.043 | 0.963 ± 0.011 |
5.345 ± 0.024 | 5.464 ± 0.027 |
3.546 ± 0.02 | 7.893 ± 0.032 |
2.272 ± 0.016 | 4.738 ± 0.024 |
3.242 ± 0.025 | 11.803 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.219 ± 0.017 | 3.095 ± 0.02 |
4.808 ± 0.025 | 4.974 ± 0.024 |
6.573 ± 0.032 | 6.031 ± 0.025 |
4.867 ± 0.035 | 7.085 ± 0.029 |
1.464 ± 0.013 | 2.525 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
