
Hubei tombus-like virus 40
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.74
Get precalculated fractions of proteins
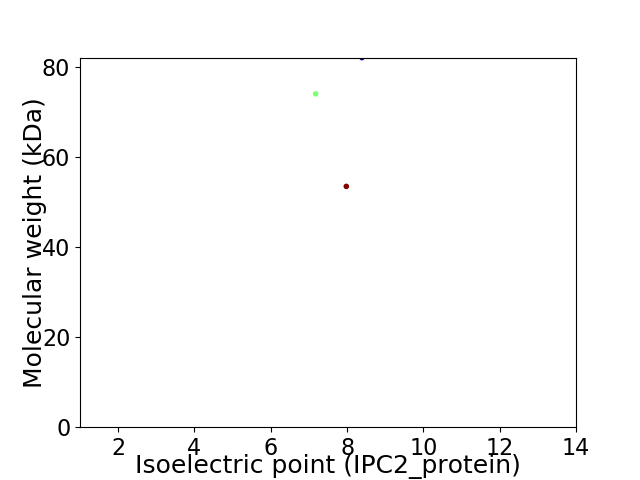
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG08|A0A1L3KG08_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 40 OX=1923289 PE=4 SV=1
MM1 pKa = 7.71PALTSDD7 pKa = 4.93LVTAPAKK14 pKa = 10.35SLQNMITGLEE24 pKa = 4.05TRR26 pKa = 11.84VLNLVPKK33 pKa = 9.41TSPLLAPSDD42 pKa = 3.76EE43 pKa = 4.18VAIIVVAVEE52 pKa = 4.35EE53 pKa = 4.39EE54 pKa = 4.37GHH56 pKa = 6.23PLSYY60 pKa = 11.01DD61 pKa = 2.98NHH63 pKa = 8.06LDD65 pKa = 3.52VLPCLGYY72 pKa = 10.4RR73 pKa = 11.84VIPMDD78 pKa = 3.86RR79 pKa = 11.84ALPPPRR85 pKa = 11.84PFEE88 pKa = 4.22TMVDD92 pKa = 4.04LQPVMDD98 pKa = 4.31RR99 pKa = 11.84YY100 pKa = 10.13EE101 pKa = 3.74RR102 pKa = 11.84QIRR105 pKa = 11.84YY106 pKa = 6.19NQKK109 pKa = 10.19RR110 pKa = 11.84GWKK113 pKa = 8.06TVILYY118 pKa = 10.63RR119 pKa = 11.84NGILLPGVYY128 pKa = 9.45PRR130 pKa = 11.84EE131 pKa = 4.13TGCIWRR137 pKa = 11.84LRR139 pKa = 11.84RR140 pKa = 11.84TIDD143 pKa = 3.32NGVEE147 pKa = 4.05CYY149 pKa = 7.78YY150 pKa = 8.9TTVARR155 pKa = 11.84SNSTIVRR162 pKa = 11.84HH163 pKa = 5.73PLINMQVEE171 pKa = 4.9PYY173 pKa = 8.99STEE176 pKa = 3.7LQPPVAGLHH185 pKa = 6.12GEE187 pKa = 4.13LSKK190 pKa = 10.71VTVTMPCRR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 10.62DD201 pKa = 3.72NPLKK205 pKa = 10.46AGARR209 pKa = 11.84EE210 pKa = 4.01FVRR213 pKa = 11.84TEE215 pKa = 3.51EE216 pKa = 4.19MAWLGDD222 pKa = 3.5VVAPVPFNEE231 pKa = 3.89WVKK234 pKa = 10.52RR235 pKa = 11.84YY236 pKa = 8.28PLKK239 pKa = 10.65RR240 pKa = 11.84RR241 pKa = 11.84EE242 pKa = 4.21QLSEE246 pKa = 3.61AHH248 pKa = 7.16DD249 pKa = 3.95RR250 pKa = 11.84LLQLGMLGTRR260 pKa = 11.84EE261 pKa = 4.24SIVKK265 pKa = 9.96NFLKK269 pKa = 10.4IEE271 pKa = 4.2TTTKK275 pKa = 10.75GVDD278 pKa = 3.45CRR280 pKa = 11.84NISPRR285 pKa = 11.84SDD287 pKa = 2.95VFLACVGPYY296 pKa = 9.32IAAVEE301 pKa = 4.25HH302 pKa = 6.45EE303 pKa = 4.51ALNAPFLVKK312 pKa = 10.25GRR314 pKa = 11.84NIRR317 pKa = 11.84ARR319 pKa = 11.84DD320 pKa = 3.1KK321 pKa = 11.13WMGSLSRR328 pKa = 11.84FSHH331 pKa = 7.18FIEE334 pKa = 5.07IDD336 pKa = 3.38FARR339 pKa = 11.84FDD341 pKa = 3.55MTLSKK346 pKa = 10.72EE347 pKa = 3.8LLEE350 pKa = 4.42NVEE353 pKa = 4.6HH354 pKa = 6.8EE355 pKa = 4.32LLLRR359 pKa = 11.84PFSEE363 pKa = 4.35DD364 pKa = 2.9QHH366 pKa = 8.33ALFHH370 pKa = 7.17IFMKK374 pKa = 10.15MALRR378 pKa = 11.84TKK380 pKa = 10.17GVSEE384 pKa = 4.78FGTSYY389 pKa = 10.51RR390 pKa = 11.84ISGTRR395 pKa = 11.84CSGDD399 pKa = 2.7AHH401 pKa = 5.81TSIGNGLLNRR411 pKa = 11.84FAIWWCLRR419 pKa = 11.84YY420 pKa = 9.63IPEE423 pKa = 3.93QDD425 pKa = 2.36WVSFHH430 pKa = 6.9EE431 pKa = 4.98GDD433 pKa = 3.99DD434 pKa = 3.99GVIGLVEE441 pKa = 4.03GRR443 pKa = 11.84VDD445 pKa = 3.42EE446 pKa = 4.79ARR448 pKa = 11.84AALEE452 pKa = 4.14SLWCLGLQAKK462 pKa = 9.73IDD464 pKa = 3.95VYY466 pKa = 11.28HH467 pKa = 7.32DD468 pKa = 4.07LSQTSFCGRR477 pKa = 11.84FMADD481 pKa = 2.95TASGMMSYY489 pKa = 10.72CDD491 pKa = 4.15PLRR494 pKa = 11.84ALAKK498 pKa = 10.15FHH500 pKa = 5.87TTVSDD505 pKa = 3.97GDD507 pKa = 3.97PKK509 pKa = 11.62ALMLAKK515 pKa = 10.31ALSYY519 pKa = 11.26YY520 pKa = 9.86STDD523 pKa = 2.96RR524 pKa = 11.84NTPVVGVLCYY534 pKa = 10.9ALITILRR541 pKa = 11.84PEE543 pKa = 4.06VTAKK547 pKa = 10.5RR548 pKa = 11.84LRR550 pKa = 11.84KK551 pKa = 9.4ALRR554 pKa = 11.84FNARR558 pKa = 11.84NLSWFAQYY566 pKa = 10.88EE567 pKa = 4.14DD568 pKa = 6.01AEE570 pKa = 5.09DD571 pKa = 3.88FWQYY575 pKa = 7.37WRR577 pKa = 11.84RR578 pKa = 11.84GVTRR582 pKa = 11.84PCCDD586 pKa = 2.68ILRR589 pKa = 11.84PFFALRR595 pKa = 11.84SGVSIRR601 pKa = 11.84EE602 pKa = 4.07QQSLEE607 pKa = 3.81QMFDD611 pKa = 2.55NWVSVGYY618 pKa = 10.73VPGEE622 pKa = 3.84FGKK625 pKa = 10.47IPVDD629 pKa = 3.26WTPLAEE635 pKa = 4.55HH636 pKa = 6.65KK637 pKa = 9.54WVTMRR642 pKa = 11.84TSDD645 pKa = 2.84WVTT648 pKa = 2.92
MM1 pKa = 7.71PALTSDD7 pKa = 4.93LVTAPAKK14 pKa = 10.35SLQNMITGLEE24 pKa = 4.05TRR26 pKa = 11.84VLNLVPKK33 pKa = 9.41TSPLLAPSDD42 pKa = 3.76EE43 pKa = 4.18VAIIVVAVEE52 pKa = 4.35EE53 pKa = 4.39EE54 pKa = 4.37GHH56 pKa = 6.23PLSYY60 pKa = 11.01DD61 pKa = 2.98NHH63 pKa = 8.06LDD65 pKa = 3.52VLPCLGYY72 pKa = 10.4RR73 pKa = 11.84VIPMDD78 pKa = 3.86RR79 pKa = 11.84ALPPPRR85 pKa = 11.84PFEE88 pKa = 4.22TMVDD92 pKa = 4.04LQPVMDD98 pKa = 4.31RR99 pKa = 11.84YY100 pKa = 10.13EE101 pKa = 3.74RR102 pKa = 11.84QIRR105 pKa = 11.84YY106 pKa = 6.19NQKK109 pKa = 10.19RR110 pKa = 11.84GWKK113 pKa = 8.06TVILYY118 pKa = 10.63RR119 pKa = 11.84NGILLPGVYY128 pKa = 9.45PRR130 pKa = 11.84EE131 pKa = 4.13TGCIWRR137 pKa = 11.84LRR139 pKa = 11.84RR140 pKa = 11.84TIDD143 pKa = 3.32NGVEE147 pKa = 4.05CYY149 pKa = 7.78YY150 pKa = 8.9TTVARR155 pKa = 11.84SNSTIVRR162 pKa = 11.84HH163 pKa = 5.73PLINMQVEE171 pKa = 4.9PYY173 pKa = 8.99STEE176 pKa = 3.7LQPPVAGLHH185 pKa = 6.12GEE187 pKa = 4.13LSKK190 pKa = 10.71VTVTMPCRR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 10.62DD201 pKa = 3.72NPLKK205 pKa = 10.46AGARR209 pKa = 11.84EE210 pKa = 4.01FVRR213 pKa = 11.84TEE215 pKa = 3.51EE216 pKa = 4.19MAWLGDD222 pKa = 3.5VVAPVPFNEE231 pKa = 3.89WVKK234 pKa = 10.52RR235 pKa = 11.84YY236 pKa = 8.28PLKK239 pKa = 10.65RR240 pKa = 11.84RR241 pKa = 11.84EE242 pKa = 4.21QLSEE246 pKa = 3.61AHH248 pKa = 7.16DD249 pKa = 3.95RR250 pKa = 11.84LLQLGMLGTRR260 pKa = 11.84EE261 pKa = 4.24SIVKK265 pKa = 9.96NFLKK269 pKa = 10.4IEE271 pKa = 4.2TTTKK275 pKa = 10.75GVDD278 pKa = 3.45CRR280 pKa = 11.84NISPRR285 pKa = 11.84SDD287 pKa = 2.95VFLACVGPYY296 pKa = 9.32IAAVEE301 pKa = 4.25HH302 pKa = 6.45EE303 pKa = 4.51ALNAPFLVKK312 pKa = 10.25GRR314 pKa = 11.84NIRR317 pKa = 11.84ARR319 pKa = 11.84DD320 pKa = 3.1KK321 pKa = 11.13WMGSLSRR328 pKa = 11.84FSHH331 pKa = 7.18FIEE334 pKa = 5.07IDD336 pKa = 3.38FARR339 pKa = 11.84FDD341 pKa = 3.55MTLSKK346 pKa = 10.72EE347 pKa = 3.8LLEE350 pKa = 4.42NVEE353 pKa = 4.6HH354 pKa = 6.8EE355 pKa = 4.32LLLRR359 pKa = 11.84PFSEE363 pKa = 4.35DD364 pKa = 2.9QHH366 pKa = 8.33ALFHH370 pKa = 7.17IFMKK374 pKa = 10.15MALRR378 pKa = 11.84TKK380 pKa = 10.17GVSEE384 pKa = 4.78FGTSYY389 pKa = 10.51RR390 pKa = 11.84ISGTRR395 pKa = 11.84CSGDD399 pKa = 2.7AHH401 pKa = 5.81TSIGNGLLNRR411 pKa = 11.84FAIWWCLRR419 pKa = 11.84YY420 pKa = 9.63IPEE423 pKa = 3.93QDD425 pKa = 2.36WVSFHH430 pKa = 6.9EE431 pKa = 4.98GDD433 pKa = 3.99DD434 pKa = 3.99GVIGLVEE441 pKa = 4.03GRR443 pKa = 11.84VDD445 pKa = 3.42EE446 pKa = 4.79ARR448 pKa = 11.84AALEE452 pKa = 4.14SLWCLGLQAKK462 pKa = 9.73IDD464 pKa = 3.95VYY466 pKa = 11.28HH467 pKa = 7.32DD468 pKa = 4.07LSQTSFCGRR477 pKa = 11.84FMADD481 pKa = 2.95TASGMMSYY489 pKa = 10.72CDD491 pKa = 4.15PLRR494 pKa = 11.84ALAKK498 pKa = 10.15FHH500 pKa = 5.87TTVSDD505 pKa = 3.97GDD507 pKa = 3.97PKK509 pKa = 11.62ALMLAKK515 pKa = 10.31ALSYY519 pKa = 11.26YY520 pKa = 9.86STDD523 pKa = 2.96RR524 pKa = 11.84NTPVVGVLCYY534 pKa = 10.9ALITILRR541 pKa = 11.84PEE543 pKa = 4.06VTAKK547 pKa = 10.5RR548 pKa = 11.84LRR550 pKa = 11.84KK551 pKa = 9.4ALRR554 pKa = 11.84FNARR558 pKa = 11.84NLSWFAQYY566 pKa = 10.88EE567 pKa = 4.14DD568 pKa = 6.01AEE570 pKa = 5.09DD571 pKa = 3.88FWQYY575 pKa = 7.37WRR577 pKa = 11.84RR578 pKa = 11.84GVTRR582 pKa = 11.84PCCDD586 pKa = 2.68ILRR589 pKa = 11.84PFFALRR595 pKa = 11.84SGVSIRR601 pKa = 11.84EE602 pKa = 4.07QQSLEE607 pKa = 3.81QMFDD611 pKa = 2.55NWVSVGYY618 pKa = 10.73VPGEE622 pKa = 3.84FGKK625 pKa = 10.47IPVDD629 pKa = 3.26WTPLAEE635 pKa = 4.55HH636 pKa = 6.65KK637 pKa = 9.54WVTMRR642 pKa = 11.84TSDD645 pKa = 2.84WVTT648 pKa = 2.92
Molecular weight: 73.95 kDa
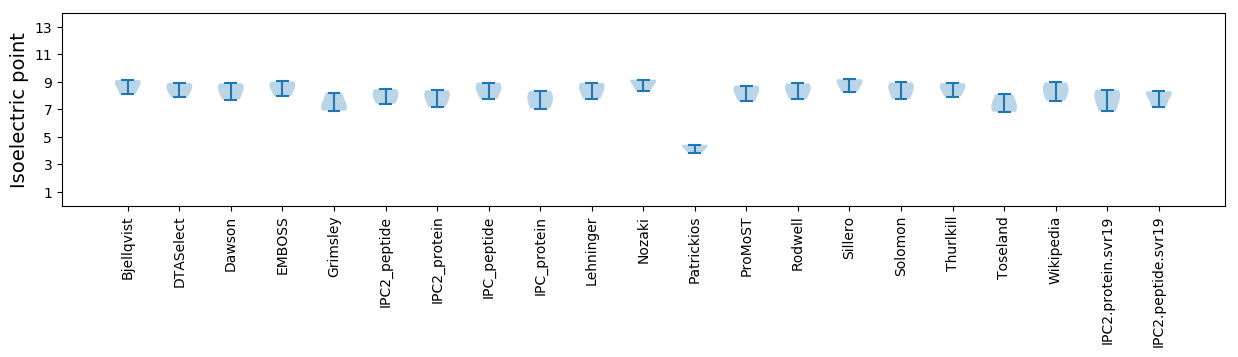
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFY2|A0A1L3KFY2_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 40 OX=1923289 PE=4 SV=1
MM1 pKa = 7.42LSYY4 pKa = 11.47YY5 pKa = 9.47EE6 pKa = 4.33GRR8 pKa = 11.84WIVLISVGVCIMVHH22 pKa = 6.32AEE24 pKa = 3.86YY25 pKa = 10.95VSAVWAVAGYY35 pKa = 8.03TVMGYY40 pKa = 8.12TYY42 pKa = 10.87SVARR46 pKa = 11.84TWGRR50 pKa = 11.84YY51 pKa = 5.24VVRR54 pKa = 11.84VRR56 pKa = 11.84SGTQFGRR63 pKa = 11.84WNEE66 pKa = 3.76IADD69 pKa = 3.68MFRR72 pKa = 11.84QYY74 pKa = 11.33VGTLDD79 pKa = 4.33RR80 pKa = 11.84PVHH83 pKa = 4.64QHH85 pKa = 4.02GHH87 pKa = 5.59HH88 pKa = 7.08NILAWEE94 pKa = 4.03RR95 pKa = 11.84RR96 pKa = 11.84VAEE99 pKa = 4.03EE100 pKa = 3.86FVVNVYY106 pKa = 10.77LGVQEE111 pKa = 5.04RR112 pKa = 11.84IRR114 pKa = 11.84DD115 pKa = 3.67VGGSRR120 pKa = 11.84TRR122 pKa = 11.84HH123 pKa = 5.51RR124 pKa = 11.84ALGDD128 pKa = 3.44RR129 pKa = 11.84KK130 pKa = 9.83HH131 pKa = 6.12VCGPTLQALDD141 pKa = 3.81NMRR144 pKa = 11.84EE145 pKa = 4.16DD146 pKa = 3.82KK147 pKa = 11.19DD148 pKa = 3.63RR149 pKa = 11.84DD150 pKa = 3.72AVFSNCRR157 pKa = 11.84KK158 pKa = 10.3SGGDD162 pKa = 3.36CPLRR166 pKa = 11.84EE167 pKa = 4.96LIPVAMLSHH176 pKa = 6.59TDD178 pKa = 3.58YY179 pKa = 11.36YY180 pKa = 9.8CTNEE184 pKa = 4.08EE185 pKa = 4.06LCSIVTGPTFIINHH199 pKa = 6.63DD200 pKa = 3.81FDD202 pKa = 4.02STTEE206 pKa = 3.93LGVYY210 pKa = 9.62EE211 pKa = 5.64DD212 pKa = 4.86EE213 pKa = 5.47DD214 pKa = 3.78EE215 pKa = 4.41QRR217 pKa = 11.84CEE219 pKa = 3.74ARR221 pKa = 11.84ATITGGQVTMQPEE234 pKa = 4.33AGSPYY239 pKa = 10.23GPHH242 pKa = 7.55RR243 pKa = 11.84YY244 pKa = 9.08HH245 pKa = 6.46LWKK248 pKa = 10.68SEE250 pKa = 3.8GVVVSRR256 pKa = 11.84TGAFTYY262 pKa = 10.38RR263 pKa = 11.84KK264 pKa = 9.48LGTIGSTTVLLGYY277 pKa = 8.41PCRR280 pKa = 11.84GIYY283 pKa = 10.23RR284 pKa = 11.84SDD286 pKa = 4.43DD287 pKa = 3.48SCQLAFADD295 pKa = 3.7QRR297 pKa = 11.84AGARR301 pKa = 11.84LSEE304 pKa = 4.29RR305 pKa = 11.84GDD307 pKa = 3.53VYY309 pKa = 11.42GKK311 pKa = 10.87GGMYY315 pKa = 10.46YY316 pKa = 10.59VDD318 pKa = 3.97GEE320 pKa = 4.42NIIFTEE326 pKa = 4.25TSDD329 pKa = 3.73RR330 pKa = 11.84LGPTGDD336 pKa = 4.07ADD338 pKa = 4.3CLPLAVLEE346 pKa = 4.64GVALQLATANRR357 pKa = 11.84DD358 pKa = 3.41EE359 pKa = 5.26KK360 pKa = 11.16YY361 pKa = 10.29LSHH364 pKa = 7.25LISYY368 pKa = 7.55TTAKK372 pKa = 10.4LHH374 pKa = 5.33AQRR377 pKa = 11.84IRR379 pKa = 11.84FRR381 pKa = 11.84RR382 pKa = 11.84GEE384 pKa = 4.23AIARR388 pKa = 11.84WVAKK392 pKa = 10.3RR393 pKa = 11.84ADD395 pKa = 3.92EE396 pKa = 4.25LALGLVQNCSLPIGDD411 pKa = 5.7PIHH414 pKa = 6.9ISTFGVIYY422 pKa = 10.0YY423 pKa = 9.95RR424 pKa = 11.84VIRR427 pKa = 11.84WVLDD431 pKa = 3.3RR432 pKa = 11.84LNKK435 pKa = 10.15FNVAEE440 pKa = 4.05YY441 pKa = 10.72LHH443 pKa = 6.39EE444 pKa = 4.45CLYY447 pKa = 11.38KK448 pKa = 10.78NLSTHH453 pKa = 5.91YY454 pKa = 9.82ALPHH458 pKa = 4.93MWVARR463 pKa = 11.84EE464 pKa = 3.72IPAYY468 pKa = 9.84NCEE471 pKa = 3.49VDD473 pKa = 4.26AIRR476 pKa = 11.84VRR478 pKa = 11.84LGRR481 pKa = 11.84LPRR484 pKa = 11.84VNRR487 pKa = 11.84PFRR490 pKa = 11.84HH491 pKa = 5.75EE492 pKa = 4.28GPRR495 pKa = 11.84ADD497 pKa = 3.28ARR499 pKa = 11.84ANIGPGHH506 pKa = 7.28RR507 pKa = 11.84PRR509 pKa = 11.84QEE511 pKa = 3.75PAEE514 pKa = 4.36HH515 pKa = 7.54DD516 pKa = 3.29NGARR520 pKa = 11.84NARR523 pKa = 11.84PEE525 pKa = 4.32FGAQNQPPAGAFGRR539 pKa = 11.84GGNYY543 pKa = 9.52RR544 pKa = 11.84GRR546 pKa = 11.84GRR548 pKa = 11.84GRR550 pKa = 11.84GAPAVVRR557 pKa = 11.84QPFRR561 pKa = 11.84RR562 pKa = 11.84PALPRR567 pKa = 11.84IPRR570 pKa = 11.84NPNGPRR576 pKa = 11.84VAPAEE581 pKa = 3.98AVRR584 pKa = 11.84DD585 pKa = 4.18DD586 pKa = 4.39GGPAAGDD593 pKa = 3.77GQIRR597 pKa = 11.84AADD600 pKa = 3.49PVQPEE605 pKa = 4.24EE606 pKa = 4.17GVEE609 pKa = 4.26DD610 pKa = 4.38GDD612 pKa = 5.97IVPEE616 pKa = 3.91WDD618 pKa = 3.35LVAWSIPEE626 pKa = 3.75RR627 pKa = 11.84DD628 pKa = 3.37RR629 pKa = 11.84LYY631 pKa = 11.44LEE633 pKa = 3.94VAEE636 pKa = 5.3NDD638 pKa = 3.5RR639 pKa = 11.84QRR641 pKa = 11.84CRR643 pKa = 11.84VLLHH647 pKa = 6.08NRR649 pKa = 11.84RR650 pKa = 11.84QVEE653 pKa = 4.72FYY655 pKa = 11.23NRR657 pKa = 11.84AAPFDD662 pKa = 3.72QHH664 pKa = 7.18AGRR667 pKa = 11.84ALLHH671 pKa = 6.28RR672 pKa = 11.84VATTRR677 pKa = 11.84GRR679 pKa = 11.84ITRR682 pKa = 11.84RR683 pKa = 11.84TIEE686 pKa = 4.74GYY688 pKa = 10.91CNDD691 pKa = 3.56ALQEE695 pKa = 4.02EE696 pKa = 4.68RR697 pKa = 11.84QPAQGRR703 pKa = 11.84GQGVRR708 pKa = 11.84PHH710 pKa = 6.9RR711 pKa = 11.84GDD713 pKa = 3.21GVVRR717 pKa = 11.84GRR719 pKa = 11.84GRR721 pKa = 11.84ARR723 pKa = 11.84PFQRR727 pKa = 11.84VGG729 pKa = 2.94
MM1 pKa = 7.42LSYY4 pKa = 11.47YY5 pKa = 9.47EE6 pKa = 4.33GRR8 pKa = 11.84WIVLISVGVCIMVHH22 pKa = 6.32AEE24 pKa = 3.86YY25 pKa = 10.95VSAVWAVAGYY35 pKa = 8.03TVMGYY40 pKa = 8.12TYY42 pKa = 10.87SVARR46 pKa = 11.84TWGRR50 pKa = 11.84YY51 pKa = 5.24VVRR54 pKa = 11.84VRR56 pKa = 11.84SGTQFGRR63 pKa = 11.84WNEE66 pKa = 3.76IADD69 pKa = 3.68MFRR72 pKa = 11.84QYY74 pKa = 11.33VGTLDD79 pKa = 4.33RR80 pKa = 11.84PVHH83 pKa = 4.64QHH85 pKa = 4.02GHH87 pKa = 5.59HH88 pKa = 7.08NILAWEE94 pKa = 4.03RR95 pKa = 11.84RR96 pKa = 11.84VAEE99 pKa = 4.03EE100 pKa = 3.86FVVNVYY106 pKa = 10.77LGVQEE111 pKa = 5.04RR112 pKa = 11.84IRR114 pKa = 11.84DD115 pKa = 3.67VGGSRR120 pKa = 11.84TRR122 pKa = 11.84HH123 pKa = 5.51RR124 pKa = 11.84ALGDD128 pKa = 3.44RR129 pKa = 11.84KK130 pKa = 9.83HH131 pKa = 6.12VCGPTLQALDD141 pKa = 3.81NMRR144 pKa = 11.84EE145 pKa = 4.16DD146 pKa = 3.82KK147 pKa = 11.19DD148 pKa = 3.63RR149 pKa = 11.84DD150 pKa = 3.72AVFSNCRR157 pKa = 11.84KK158 pKa = 10.3SGGDD162 pKa = 3.36CPLRR166 pKa = 11.84EE167 pKa = 4.96LIPVAMLSHH176 pKa = 6.59TDD178 pKa = 3.58YY179 pKa = 11.36YY180 pKa = 9.8CTNEE184 pKa = 4.08EE185 pKa = 4.06LCSIVTGPTFIINHH199 pKa = 6.63DD200 pKa = 3.81FDD202 pKa = 4.02STTEE206 pKa = 3.93LGVYY210 pKa = 9.62EE211 pKa = 5.64DD212 pKa = 4.86EE213 pKa = 5.47DD214 pKa = 3.78EE215 pKa = 4.41QRR217 pKa = 11.84CEE219 pKa = 3.74ARR221 pKa = 11.84ATITGGQVTMQPEE234 pKa = 4.33AGSPYY239 pKa = 10.23GPHH242 pKa = 7.55RR243 pKa = 11.84YY244 pKa = 9.08HH245 pKa = 6.46LWKK248 pKa = 10.68SEE250 pKa = 3.8GVVVSRR256 pKa = 11.84TGAFTYY262 pKa = 10.38RR263 pKa = 11.84KK264 pKa = 9.48LGTIGSTTVLLGYY277 pKa = 8.41PCRR280 pKa = 11.84GIYY283 pKa = 10.23RR284 pKa = 11.84SDD286 pKa = 4.43DD287 pKa = 3.48SCQLAFADD295 pKa = 3.7QRR297 pKa = 11.84AGARR301 pKa = 11.84LSEE304 pKa = 4.29RR305 pKa = 11.84GDD307 pKa = 3.53VYY309 pKa = 11.42GKK311 pKa = 10.87GGMYY315 pKa = 10.46YY316 pKa = 10.59VDD318 pKa = 3.97GEE320 pKa = 4.42NIIFTEE326 pKa = 4.25TSDD329 pKa = 3.73RR330 pKa = 11.84LGPTGDD336 pKa = 4.07ADD338 pKa = 4.3CLPLAVLEE346 pKa = 4.64GVALQLATANRR357 pKa = 11.84DD358 pKa = 3.41EE359 pKa = 5.26KK360 pKa = 11.16YY361 pKa = 10.29LSHH364 pKa = 7.25LISYY368 pKa = 7.55TTAKK372 pKa = 10.4LHH374 pKa = 5.33AQRR377 pKa = 11.84IRR379 pKa = 11.84FRR381 pKa = 11.84RR382 pKa = 11.84GEE384 pKa = 4.23AIARR388 pKa = 11.84WVAKK392 pKa = 10.3RR393 pKa = 11.84ADD395 pKa = 3.92EE396 pKa = 4.25LALGLVQNCSLPIGDD411 pKa = 5.7PIHH414 pKa = 6.9ISTFGVIYY422 pKa = 10.0YY423 pKa = 9.95RR424 pKa = 11.84VIRR427 pKa = 11.84WVLDD431 pKa = 3.3RR432 pKa = 11.84LNKK435 pKa = 10.15FNVAEE440 pKa = 4.05YY441 pKa = 10.72LHH443 pKa = 6.39EE444 pKa = 4.45CLYY447 pKa = 11.38KK448 pKa = 10.78NLSTHH453 pKa = 5.91YY454 pKa = 9.82ALPHH458 pKa = 4.93MWVARR463 pKa = 11.84EE464 pKa = 3.72IPAYY468 pKa = 9.84NCEE471 pKa = 3.49VDD473 pKa = 4.26AIRR476 pKa = 11.84VRR478 pKa = 11.84LGRR481 pKa = 11.84LPRR484 pKa = 11.84VNRR487 pKa = 11.84PFRR490 pKa = 11.84HH491 pKa = 5.75EE492 pKa = 4.28GPRR495 pKa = 11.84ADD497 pKa = 3.28ARR499 pKa = 11.84ANIGPGHH506 pKa = 7.28RR507 pKa = 11.84PRR509 pKa = 11.84QEE511 pKa = 3.75PAEE514 pKa = 4.36HH515 pKa = 7.54DD516 pKa = 3.29NGARR520 pKa = 11.84NARR523 pKa = 11.84PEE525 pKa = 4.32FGAQNQPPAGAFGRR539 pKa = 11.84GGNYY543 pKa = 9.52RR544 pKa = 11.84GRR546 pKa = 11.84GRR548 pKa = 11.84GRR550 pKa = 11.84GAPAVVRR557 pKa = 11.84QPFRR561 pKa = 11.84RR562 pKa = 11.84PALPRR567 pKa = 11.84IPRR570 pKa = 11.84NPNGPRR576 pKa = 11.84VAPAEE581 pKa = 3.98AVRR584 pKa = 11.84DD585 pKa = 4.18DD586 pKa = 4.39GGPAAGDD593 pKa = 3.77GQIRR597 pKa = 11.84AADD600 pKa = 3.49PVQPEE605 pKa = 4.24EE606 pKa = 4.17GVEE609 pKa = 4.26DD610 pKa = 4.38GDD612 pKa = 5.97IVPEE616 pKa = 3.91WDD618 pKa = 3.35LVAWSIPEE626 pKa = 3.75RR627 pKa = 11.84DD628 pKa = 3.37RR629 pKa = 11.84LYY631 pKa = 11.44LEE633 pKa = 3.94VAEE636 pKa = 5.3NDD638 pKa = 3.5RR639 pKa = 11.84QRR641 pKa = 11.84CRR643 pKa = 11.84VLLHH647 pKa = 6.08NRR649 pKa = 11.84RR650 pKa = 11.84QVEE653 pKa = 4.72FYY655 pKa = 11.23NRR657 pKa = 11.84AAPFDD662 pKa = 3.72QHH664 pKa = 7.18AGRR667 pKa = 11.84ALLHH671 pKa = 6.28RR672 pKa = 11.84VATTRR677 pKa = 11.84GRR679 pKa = 11.84ITRR682 pKa = 11.84RR683 pKa = 11.84TIEE686 pKa = 4.74GYY688 pKa = 10.91CNDD691 pKa = 3.56ALQEE695 pKa = 4.02EE696 pKa = 4.68RR697 pKa = 11.84QPAQGRR703 pKa = 11.84GQGVRR708 pKa = 11.84PHH710 pKa = 6.9RR711 pKa = 11.84GDD713 pKa = 3.21GVVRR717 pKa = 11.84GRR719 pKa = 11.84GRR721 pKa = 11.84ARR723 pKa = 11.84PFQRR727 pKa = 11.84VGG729 pKa = 2.94
Molecular weight: 81.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1857 |
480 |
729 |
619.0 |
69.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.539 ± 0.774 | 2.423 ± 0.301 |
4.685 ± 0.787 | 5.924 ± 0.294 |
2.854 ± 0.475 | 7.593 ± 1.219 |
2.423 ± 0.492 | 4.631 ± 0.144 |
3.016 ± 0.764 | 9.532 ± 1.506 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.477 ± 0.652 | 3.069 ± 0.345 |
5.439 ± 0.272 | 3.285 ± 0.273 |
9.155 ± 1.726 | 6.354 ± 1.871 |
6.139 ± 1.032 | 7.324 ± 0.708 |
2.208 ± 0.366 | 3.931 ± 0.289 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
