
Asparagus virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Alphaflexiviridae; Potexvirus
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
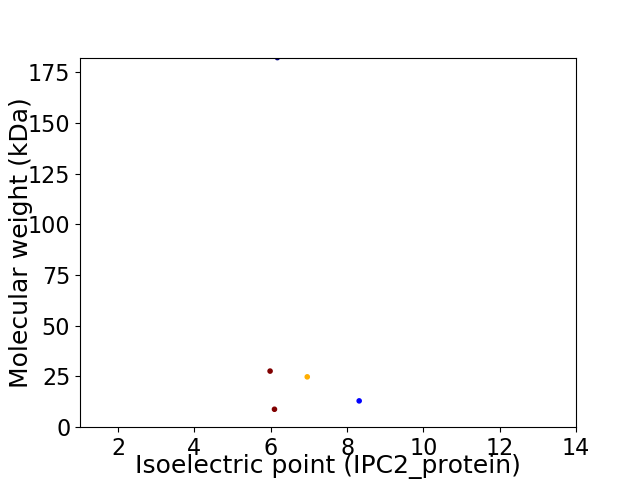
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B1B3P0|B1B3P0_9VIRU Triple gene block protein 2 OS=Asparagus virus 3 OX=445435 GN=TGBp2 PE=4 SV=1
MM1 pKa = 7.63EE2 pKa = 5.13ISYY5 pKa = 10.51IVDD8 pKa = 4.1LLNFLGFTRR17 pKa = 11.84SSRR20 pKa = 11.84PFSLPLVVHH29 pKa = 6.42GVAGSGKK36 pKa = 7.04THH38 pKa = 7.55LLRR41 pKa = 11.84KK42 pKa = 9.73VSLHH46 pKa = 5.51FPEE49 pKa = 5.79LVHH52 pKa = 7.6CSFTPQLIDD61 pKa = 3.67PNSGRR66 pKa = 11.84RR67 pKa = 11.84QLPASEE73 pKa = 4.61TPTDD77 pKa = 4.68LLDD80 pKa = 4.34EE81 pKa = 4.43YY82 pKa = 11.17LGGPNPVVRR91 pKa = 11.84LLKK94 pKa = 10.82VCDD97 pKa = 3.82PLQYY101 pKa = 10.71NCPDD105 pKa = 3.39PEE107 pKa = 4.64IPHH110 pKa = 5.8FQSLTTRR117 pKa = 11.84RR118 pKa = 11.84FCPLTTTLLNSLFGTNIVSAVPTCCRR144 pKa = 11.84IEE146 pKa = 3.98IQDD149 pKa = 4.38PYY151 pKa = 10.33STDD154 pKa = 3.95PIGTVVTFSPEE165 pKa = 3.14IHH167 pKa = 6.36TLLSRR172 pKa = 11.84HH173 pKa = 5.76GCQPTPISEE182 pKa = 4.08LWGLNIRR189 pKa = 11.84VVSCYY194 pKa = 10.34VDD196 pKa = 3.39SLEE199 pKa = 4.54EE200 pKa = 3.69ALLNHH205 pKa = 7.23RR206 pKa = 11.84APLFLALTRR215 pKa = 11.84HH216 pKa = 4.98TAEE219 pKa = 3.64LHH221 pKa = 6.05IFLFDD226 pKa = 3.34ARR228 pKa = 11.84TDD230 pKa = 3.18AAYY233 pKa = 9.98EE234 pKa = 3.95LRR236 pKa = 11.84KK237 pKa = 9.9CLQDD241 pKa = 3.24PRR243 pKa = 11.84HH244 pKa = 6.27RR245 pKa = 11.84GSS247 pKa = 3.03
MM1 pKa = 7.63EE2 pKa = 5.13ISYY5 pKa = 10.51IVDD8 pKa = 4.1LLNFLGFTRR17 pKa = 11.84SSRR20 pKa = 11.84PFSLPLVVHH29 pKa = 6.42GVAGSGKK36 pKa = 7.04THH38 pKa = 7.55LLRR41 pKa = 11.84KK42 pKa = 9.73VSLHH46 pKa = 5.51FPEE49 pKa = 5.79LVHH52 pKa = 7.6CSFTPQLIDD61 pKa = 3.67PNSGRR66 pKa = 11.84RR67 pKa = 11.84QLPASEE73 pKa = 4.61TPTDD77 pKa = 4.68LLDD80 pKa = 4.34EE81 pKa = 4.43YY82 pKa = 11.17LGGPNPVVRR91 pKa = 11.84LLKK94 pKa = 10.82VCDD97 pKa = 3.82PLQYY101 pKa = 10.71NCPDD105 pKa = 3.39PEE107 pKa = 4.64IPHH110 pKa = 5.8FQSLTTRR117 pKa = 11.84RR118 pKa = 11.84FCPLTTTLLNSLFGTNIVSAVPTCCRR144 pKa = 11.84IEE146 pKa = 3.98IQDD149 pKa = 4.38PYY151 pKa = 10.33STDD154 pKa = 3.95PIGTVVTFSPEE165 pKa = 3.14IHH167 pKa = 6.36TLLSRR172 pKa = 11.84HH173 pKa = 5.76GCQPTPISEE182 pKa = 4.08LWGLNIRR189 pKa = 11.84VVSCYY194 pKa = 10.34VDD196 pKa = 3.39SLEE199 pKa = 4.54EE200 pKa = 3.69ALLNHH205 pKa = 7.23RR206 pKa = 11.84APLFLALTRR215 pKa = 11.84HH216 pKa = 4.98TAEE219 pKa = 3.64LHH221 pKa = 6.05IFLFDD226 pKa = 3.34ARR228 pKa = 11.84TDD230 pKa = 3.18AAYY233 pKa = 9.98EE234 pKa = 3.95LRR236 pKa = 11.84KK237 pKa = 9.9CLQDD241 pKa = 3.24PRR243 pKa = 11.84HH244 pKa = 6.27RR245 pKa = 11.84GSS247 pKa = 3.03
Molecular weight: 27.65 kDa
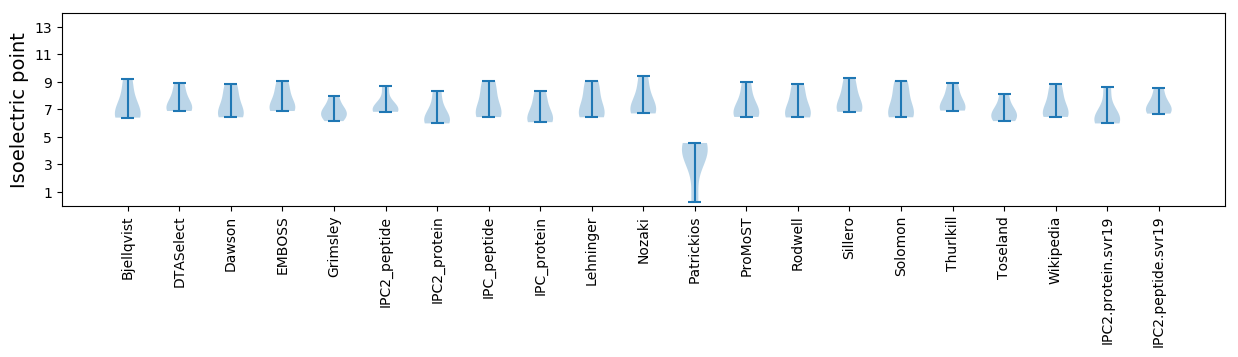
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B1B3P1|B1B3P1_9VIRU Movement protein TGBp3 OS=Asparagus virus 3 OX=445435 GN=TGBp3 PE=3 SV=1
MM1 pKa = 7.83PGLTPPTNYY10 pKa = 10.27EE11 pKa = 4.2SVFKK15 pKa = 10.64ILAIGALSCLSIYY28 pKa = 8.92SLRR31 pKa = 11.84TNHH34 pKa = 6.73LPHH37 pKa = 6.67TGDD40 pKa = 4.27NIHH43 pKa = 6.69HH44 pKa = 6.93LPHH47 pKa = 6.69GGNYY51 pKa = 10.1ADD53 pKa = 3.56GTKK56 pKa = 9.71RR57 pKa = 11.84VQYY60 pKa = 8.72FRR62 pKa = 11.84PSAPVHH68 pKa = 5.84GSSKK72 pKa = 9.13FTAACAILFLTLLILAQSQWPARR95 pKa = 11.84AVRR98 pKa = 11.84CSVRR102 pKa = 11.84VCGHH106 pKa = 6.28CHH108 pKa = 6.95PDD110 pKa = 2.64STMPSNSDD118 pKa = 2.92RR119 pKa = 3.88
MM1 pKa = 7.83PGLTPPTNYY10 pKa = 10.27EE11 pKa = 4.2SVFKK15 pKa = 10.64ILAIGALSCLSIYY28 pKa = 8.92SLRR31 pKa = 11.84TNHH34 pKa = 6.73LPHH37 pKa = 6.67TGDD40 pKa = 4.27NIHH43 pKa = 6.69HH44 pKa = 6.93LPHH47 pKa = 6.69GGNYY51 pKa = 10.1ADD53 pKa = 3.56GTKK56 pKa = 9.71RR57 pKa = 11.84VQYY60 pKa = 8.72FRR62 pKa = 11.84PSAPVHH68 pKa = 5.84GSSKK72 pKa = 9.13FTAACAILFLTLLILAQSQWPARR95 pKa = 11.84AVRR98 pKa = 11.84CSVRR102 pKa = 11.84VCGHH106 pKa = 6.28CHH108 pKa = 6.95PDD110 pKa = 2.64STMPSNSDD118 pKa = 2.92RR119 pKa = 3.88
Molecular weight: 12.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2295 |
85 |
1614 |
459.0 |
51.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.235 ± 1.045 | 1.917 ± 0.806 |
5.403 ± 0.743 | 4.88 ± 0.87 |
4.357 ± 0.19 | 4.706 ± 0.545 |
3.66 ± 0.631 | 5.142 ± 0.244 |
4.575 ± 1.197 | 10.109 ± 1.329 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.539 | 3.704 ± 0.224 |
7.625 ± 0.744 | 3.747 ± 0.355 |
5.316 ± 0.455 | 7.277 ± 0.705 |
7.495 ± 0.322 | 5.403 ± 1.02 |
1.046 ± 0.176 | 3.268 ± 0.444 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
