
Beihai weivirus-like virus 16
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.48
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLA5|A0A1L3KLA5_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 16 OX=1922744 PE=4 SV=1
MM1 pKa = 7.48HH2 pKa = 7.13SFVPRR7 pKa = 11.84PEE9 pKa = 4.55CYY11 pKa = 10.24APLCSRR17 pKa = 11.84CDD19 pKa = 3.63DD20 pKa = 3.32QSFRR24 pKa = 11.84SLQTGRR30 pKa = 11.84VEE32 pKa = 3.4IHH34 pKa = 6.11MLVTDD39 pKa = 4.0GCEE42 pKa = 3.88ACRR45 pKa = 11.84SLIPSCLRR53 pKa = 11.84EE54 pKa = 3.89FTRR57 pKa = 11.84TVRR60 pKa = 11.84TFRR63 pKa = 11.84LEE65 pKa = 3.93DD66 pKa = 3.45DD67 pKa = 4.37FFARR71 pKa = 11.84LCIDD75 pKa = 3.4YY76 pKa = 8.92STAADD81 pKa = 3.36KK82 pKa = 11.16SLNQSLTSEE91 pKa = 4.06IRR93 pKa = 11.84SEE95 pKa = 3.95KK96 pKa = 10.76DD97 pKa = 2.83PLRR100 pKa = 11.84RR101 pKa = 11.84VAMMRR106 pKa = 11.84LVPEE110 pKa = 4.62ALSQVHH116 pKa = 5.98ARR118 pKa = 11.84QRR120 pKa = 11.84LLSPRR125 pKa = 11.84FNRR128 pKa = 11.84HH129 pKa = 4.46SLFLYY134 pKa = 10.29GAGGLTTVTVLTLGTRR150 pKa = 11.84RR151 pKa = 11.84LLANWDD157 pKa = 3.3MPKK160 pKa = 10.0PVRR163 pKa = 11.84YY164 pKa = 9.7GLTAFTMFPFWEE176 pKa = 4.12MGVFFLGRR184 pKa = 11.84KK185 pKa = 8.47VSQMWSGHH193 pKa = 4.23AHH195 pKa = 5.52YY196 pKa = 10.31KK197 pKa = 9.93ILPLRR202 pKa = 11.84DD203 pKa = 3.08ITEE206 pKa = 4.35TEE208 pKa = 4.3DD209 pKa = 3.35EE210 pKa = 5.44DD211 pKa = 4.37IVEE214 pKa = 4.8DD215 pKa = 3.96PAVPAPVPRR224 pKa = 11.84IAPTLPEE231 pKa = 5.05PDD233 pKa = 4.17SDD235 pKa = 4.14HH236 pKa = 7.27SDD238 pKa = 3.44DD239 pKa = 3.98VSSVTTDD246 pKa = 3.52PGAHH250 pKa = 6.08MDD252 pKa = 4.57EE253 pKa = 4.05IRR255 pKa = 11.84NWFQNLPPPPGLGPDD270 pKa = 3.56DD271 pKa = 5.52RR272 pKa = 11.84IPVNTIVGPLEE283 pKa = 4.18SVILEE288 pKa = 4.21TNIEE292 pKa = 4.11SGEE295 pKa = 4.25DD296 pKa = 3.21YY297 pKa = 10.62LARR300 pKa = 11.84VEE302 pKa = 4.28TRR304 pKa = 11.84DD305 pKa = 3.28IPVPRR310 pKa = 11.84VATPIGPTTQEE321 pKa = 3.77VQHH324 pKa = 6.32YY325 pKa = 10.47SDD327 pKa = 3.79ALEE330 pKa = 4.39NIIRR334 pKa = 11.84ACKK337 pKa = 10.17GRR339 pKa = 11.84VEE341 pKa = 4.97GKK343 pKa = 10.15AKK345 pKa = 10.19PFKK348 pKa = 10.62PSDD351 pKa = 3.32KK352 pKa = 10.27VYY354 pKa = 11.15AGILSVQRR362 pKa = 11.84ALKK365 pKa = 10.53DD366 pKa = 3.23HH367 pKa = 6.48VFTKK371 pKa = 10.48RR372 pKa = 11.84RR373 pKa = 11.84VQQWAADD380 pKa = 3.91NPCLNEE386 pKa = 4.04LVSKK390 pKa = 10.13KK391 pKa = 8.21WSQRR395 pKa = 11.84TVVNALDD402 pKa = 3.54MLMATSANQHH412 pKa = 4.44FVKK415 pKa = 10.0WAVSIKK421 pKa = 10.92DD422 pKa = 3.61EE423 pKa = 3.92ILAAGKK429 pKa = 8.79EE430 pKa = 3.96PRR432 pKa = 11.84MIMSCGGAGQLCALLTVKK450 pKa = 10.41CFEE453 pKa = 5.05DD454 pKa = 4.72LYY456 pKa = 10.97FDD458 pKa = 3.45HH459 pKa = 7.57FEE461 pKa = 3.96RR462 pKa = 11.84RR463 pKa = 11.84SIKK466 pKa = 10.03HH467 pKa = 5.27RR468 pKa = 11.84PKK470 pKa = 8.75MVAMRR475 pKa = 11.84EE476 pKa = 4.13VAEE479 pKa = 4.1KK480 pKa = 9.63MRR482 pKa = 11.84YY483 pKa = 7.68GHH485 pKa = 6.66VYY487 pKa = 10.46EE488 pKa = 4.98NDD490 pKa = 2.96GSAWDD495 pKa = 3.79VTVSPEE501 pKa = 4.36LRR503 pKa = 11.84HH504 pKa = 5.35TLEE507 pKa = 4.85DD508 pKa = 5.13PILEE512 pKa = 4.71HH513 pKa = 7.38IAMLLFSHH521 pKa = 6.72GNFEE525 pKa = 5.82LITWDD530 pKa = 3.37MQGADD535 pKa = 3.59LEE537 pKa = 4.44DD538 pKa = 3.46RR539 pKa = 11.84RR540 pKa = 11.84KK541 pKa = 10.62DD542 pKa = 3.39QLKK545 pKa = 10.8ASFNKK550 pKa = 10.11HH551 pKa = 4.51GARR554 pKa = 11.84ACIIVRR560 pKa = 11.84AFRR563 pKa = 11.84KK564 pKa = 10.3SGDD567 pKa = 3.44RR568 pKa = 11.84GTSALNNLVNLTLWSAMVLDD588 pKa = 5.28DD589 pKa = 4.28PAQVVYY595 pKa = 10.48KK596 pKa = 9.95PSARR600 pKa = 11.84KK601 pKa = 9.54YY602 pKa = 10.44RR603 pKa = 11.84INGQNTDD610 pKa = 3.36FAFFYY615 pKa = 10.48EE616 pKa = 4.74GDD618 pKa = 3.91DD619 pKa = 4.31SIVSCGQRR627 pKa = 11.84LDD629 pKa = 3.6VDD631 pKa = 3.87HH632 pKa = 6.86LTKK635 pKa = 10.11EE636 pKa = 3.98WEE638 pKa = 4.2RR639 pKa = 11.84AGMRR643 pKa = 11.84PKK645 pKa = 10.6LFHH648 pKa = 6.82RR649 pKa = 11.84KK650 pKa = 9.66KK651 pKa = 9.62GDD653 pKa = 2.47AWTFCGVNGIAGTLAEE669 pKa = 5.03PVPQLARR676 pKa = 11.84NIASSAYY683 pKa = 10.0SVSQGLDD690 pKa = 2.84TGLARR695 pKa = 11.84AMTLVGRR702 pKa = 11.84VEE704 pKa = 4.25NFRR707 pKa = 11.84DD708 pKa = 3.54SCPVFAHH715 pKa = 5.95YY716 pKa = 8.0FACIARR722 pKa = 11.84HH723 pKa = 5.53HH724 pKa = 6.35LQGCTLKK731 pKa = 10.58SVPMDD736 pKa = 3.95RR737 pKa = 11.84EE738 pKa = 4.09SQMQIYY744 pKa = 10.19GDD746 pKa = 3.88YY747 pKa = 10.88DD748 pKa = 3.67DD749 pKa = 5.54SRR751 pKa = 11.84TVNIEE756 pKa = 3.78DD757 pKa = 4.67LLVVDD762 pKa = 5.11NRR764 pKa = 11.84TSTKK768 pKa = 9.25GTDD771 pKa = 3.6VIQAACGAPLTQEE784 pKa = 4.21LVSAIAGLDD793 pKa = 3.37TLGPEE798 pKa = 4.13DD799 pKa = 3.29TWIFRR804 pKa = 11.84HH805 pKa = 6.02MPAEE809 pKa = 4.19WFHH812 pKa = 7.48VPAA815 pKa = 5.9
MM1 pKa = 7.48HH2 pKa = 7.13SFVPRR7 pKa = 11.84PEE9 pKa = 4.55CYY11 pKa = 10.24APLCSRR17 pKa = 11.84CDD19 pKa = 3.63DD20 pKa = 3.32QSFRR24 pKa = 11.84SLQTGRR30 pKa = 11.84VEE32 pKa = 3.4IHH34 pKa = 6.11MLVTDD39 pKa = 4.0GCEE42 pKa = 3.88ACRR45 pKa = 11.84SLIPSCLRR53 pKa = 11.84EE54 pKa = 3.89FTRR57 pKa = 11.84TVRR60 pKa = 11.84TFRR63 pKa = 11.84LEE65 pKa = 3.93DD66 pKa = 3.45DD67 pKa = 4.37FFARR71 pKa = 11.84LCIDD75 pKa = 3.4YY76 pKa = 8.92STAADD81 pKa = 3.36KK82 pKa = 11.16SLNQSLTSEE91 pKa = 4.06IRR93 pKa = 11.84SEE95 pKa = 3.95KK96 pKa = 10.76DD97 pKa = 2.83PLRR100 pKa = 11.84RR101 pKa = 11.84VAMMRR106 pKa = 11.84LVPEE110 pKa = 4.62ALSQVHH116 pKa = 5.98ARR118 pKa = 11.84QRR120 pKa = 11.84LLSPRR125 pKa = 11.84FNRR128 pKa = 11.84HH129 pKa = 4.46SLFLYY134 pKa = 10.29GAGGLTTVTVLTLGTRR150 pKa = 11.84RR151 pKa = 11.84LLANWDD157 pKa = 3.3MPKK160 pKa = 10.0PVRR163 pKa = 11.84YY164 pKa = 9.7GLTAFTMFPFWEE176 pKa = 4.12MGVFFLGRR184 pKa = 11.84KK185 pKa = 8.47VSQMWSGHH193 pKa = 4.23AHH195 pKa = 5.52YY196 pKa = 10.31KK197 pKa = 9.93ILPLRR202 pKa = 11.84DD203 pKa = 3.08ITEE206 pKa = 4.35TEE208 pKa = 4.3DD209 pKa = 3.35EE210 pKa = 5.44DD211 pKa = 4.37IVEE214 pKa = 4.8DD215 pKa = 3.96PAVPAPVPRR224 pKa = 11.84IAPTLPEE231 pKa = 5.05PDD233 pKa = 4.17SDD235 pKa = 4.14HH236 pKa = 7.27SDD238 pKa = 3.44DD239 pKa = 3.98VSSVTTDD246 pKa = 3.52PGAHH250 pKa = 6.08MDD252 pKa = 4.57EE253 pKa = 4.05IRR255 pKa = 11.84NWFQNLPPPPGLGPDD270 pKa = 3.56DD271 pKa = 5.52RR272 pKa = 11.84IPVNTIVGPLEE283 pKa = 4.18SVILEE288 pKa = 4.21TNIEE292 pKa = 4.11SGEE295 pKa = 4.25DD296 pKa = 3.21YY297 pKa = 10.62LARR300 pKa = 11.84VEE302 pKa = 4.28TRR304 pKa = 11.84DD305 pKa = 3.28IPVPRR310 pKa = 11.84VATPIGPTTQEE321 pKa = 3.77VQHH324 pKa = 6.32YY325 pKa = 10.47SDD327 pKa = 3.79ALEE330 pKa = 4.39NIIRR334 pKa = 11.84ACKK337 pKa = 10.17GRR339 pKa = 11.84VEE341 pKa = 4.97GKK343 pKa = 10.15AKK345 pKa = 10.19PFKK348 pKa = 10.62PSDD351 pKa = 3.32KK352 pKa = 10.27VYY354 pKa = 11.15AGILSVQRR362 pKa = 11.84ALKK365 pKa = 10.53DD366 pKa = 3.23HH367 pKa = 6.48VFTKK371 pKa = 10.48RR372 pKa = 11.84RR373 pKa = 11.84VQQWAADD380 pKa = 3.91NPCLNEE386 pKa = 4.04LVSKK390 pKa = 10.13KK391 pKa = 8.21WSQRR395 pKa = 11.84TVVNALDD402 pKa = 3.54MLMATSANQHH412 pKa = 4.44FVKK415 pKa = 10.0WAVSIKK421 pKa = 10.92DD422 pKa = 3.61EE423 pKa = 3.92ILAAGKK429 pKa = 8.79EE430 pKa = 3.96PRR432 pKa = 11.84MIMSCGGAGQLCALLTVKK450 pKa = 10.41CFEE453 pKa = 5.05DD454 pKa = 4.72LYY456 pKa = 10.97FDD458 pKa = 3.45HH459 pKa = 7.57FEE461 pKa = 3.96RR462 pKa = 11.84RR463 pKa = 11.84SIKK466 pKa = 10.03HH467 pKa = 5.27RR468 pKa = 11.84PKK470 pKa = 8.75MVAMRR475 pKa = 11.84EE476 pKa = 4.13VAEE479 pKa = 4.1KK480 pKa = 9.63MRR482 pKa = 11.84YY483 pKa = 7.68GHH485 pKa = 6.66VYY487 pKa = 10.46EE488 pKa = 4.98NDD490 pKa = 2.96GSAWDD495 pKa = 3.79VTVSPEE501 pKa = 4.36LRR503 pKa = 11.84HH504 pKa = 5.35TLEE507 pKa = 4.85DD508 pKa = 5.13PILEE512 pKa = 4.71HH513 pKa = 7.38IAMLLFSHH521 pKa = 6.72GNFEE525 pKa = 5.82LITWDD530 pKa = 3.37MQGADD535 pKa = 3.59LEE537 pKa = 4.44DD538 pKa = 3.46RR539 pKa = 11.84RR540 pKa = 11.84KK541 pKa = 10.62DD542 pKa = 3.39QLKK545 pKa = 10.8ASFNKK550 pKa = 10.11HH551 pKa = 4.51GARR554 pKa = 11.84ACIIVRR560 pKa = 11.84AFRR563 pKa = 11.84KK564 pKa = 10.3SGDD567 pKa = 3.44RR568 pKa = 11.84GTSALNNLVNLTLWSAMVLDD588 pKa = 5.28DD589 pKa = 4.28PAQVVYY595 pKa = 10.48KK596 pKa = 9.95PSARR600 pKa = 11.84KK601 pKa = 9.54YY602 pKa = 10.44RR603 pKa = 11.84INGQNTDD610 pKa = 3.36FAFFYY615 pKa = 10.48EE616 pKa = 4.74GDD618 pKa = 3.91DD619 pKa = 4.31SIVSCGQRR627 pKa = 11.84LDD629 pKa = 3.6VDD631 pKa = 3.87HH632 pKa = 6.86LTKK635 pKa = 10.11EE636 pKa = 3.98WEE638 pKa = 4.2RR639 pKa = 11.84AGMRR643 pKa = 11.84PKK645 pKa = 10.6LFHH648 pKa = 6.82RR649 pKa = 11.84KK650 pKa = 9.66KK651 pKa = 9.62GDD653 pKa = 2.47AWTFCGVNGIAGTLAEE669 pKa = 5.03PVPQLARR676 pKa = 11.84NIASSAYY683 pKa = 10.0SVSQGLDD690 pKa = 2.84TGLARR695 pKa = 11.84AMTLVGRR702 pKa = 11.84VEE704 pKa = 4.25NFRR707 pKa = 11.84DD708 pKa = 3.54SCPVFAHH715 pKa = 5.95YY716 pKa = 8.0FACIARR722 pKa = 11.84HH723 pKa = 5.53HH724 pKa = 6.35LQGCTLKK731 pKa = 10.58SVPMDD736 pKa = 3.95RR737 pKa = 11.84EE738 pKa = 4.09SQMQIYY744 pKa = 10.19GDD746 pKa = 3.88YY747 pKa = 10.88DD748 pKa = 3.67DD749 pKa = 5.54SRR751 pKa = 11.84TVNIEE756 pKa = 3.78DD757 pKa = 4.67LLVVDD762 pKa = 5.11NRR764 pKa = 11.84TSTKK768 pKa = 9.25GTDD771 pKa = 3.6VIQAACGAPLTQEE784 pKa = 4.21LVSAIAGLDD793 pKa = 3.37TLGPEE798 pKa = 4.13DD799 pKa = 3.29TWIFRR804 pKa = 11.84HH805 pKa = 6.02MPAEE809 pKa = 4.19WFHH812 pKa = 7.48VPAA815 pKa = 5.9
Molecular weight: 91.71 kDa
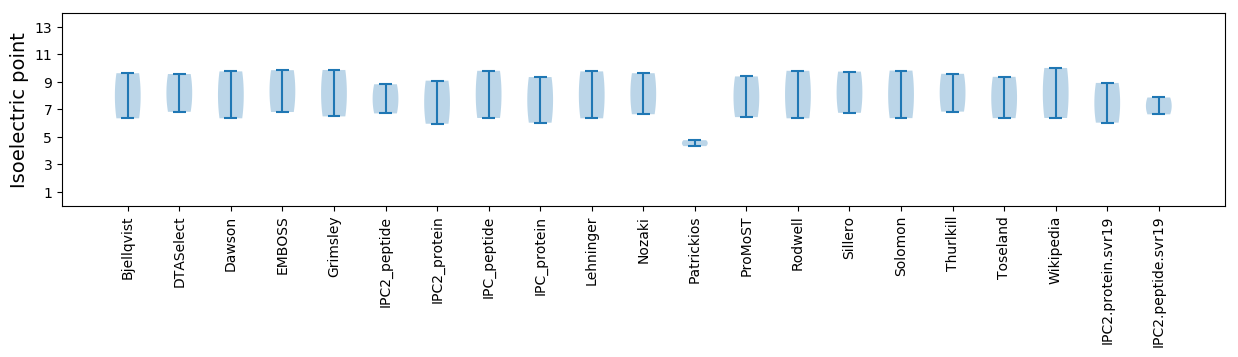
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLA5|A0A1L3KLA5_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 16 OX=1922744 PE=4 SV=1
MM1 pKa = 7.48AQRR4 pKa = 11.84KK5 pKa = 7.12SRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84NGNSQQNGQQNGQQLALRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84NGRR33 pKa = 11.84RR34 pKa = 11.84GPNARR39 pKa = 11.84TLVPGVGSTPGSPFGGIVGSDD60 pKa = 3.35LRR62 pKa = 11.84CWDD65 pKa = 3.46AKK67 pKa = 11.05LPLHH71 pKa = 6.59LPLPRR76 pKa = 11.84AVGPYY81 pKa = 9.14TVIRR85 pKa = 11.84CTRR88 pKa = 11.84RR89 pKa = 11.84INTANRR95 pKa = 11.84QMLFGTFQDD104 pKa = 4.08DD105 pKa = 3.99DD106 pKa = 4.08GTLGLVWNNICALGAINHH124 pKa = 6.83HH125 pKa = 6.85DD126 pKa = 4.91PINAANNTQRR136 pKa = 11.84YY137 pKa = 6.91TIDD140 pKa = 3.28LGGLGSGCSLVPAAMTVQVMNPNALQTTSGVIYY173 pKa = 10.15AGVMNTQAAFGGDD186 pKa = 3.03STAYY190 pKa = 10.26GDD192 pKa = 3.93RR193 pKa = 11.84FDD195 pKa = 4.76TFVEE199 pKa = 4.71FQNPRR204 pKa = 11.84LCAAPKK210 pKa = 9.52LALRR214 pKa = 11.84GVQMSSYY221 pKa = 8.83PLNMSKK227 pKa = 10.41VSEE230 pKa = 4.17FTGLGVATDD239 pKa = 4.75SVITWSDD246 pKa = 3.04PTLRR250 pKa = 11.84PTGWAPIMIYY260 pKa = 8.89NTAGTTLEE268 pKa = 3.94YY269 pKa = 10.61LVTIEE274 pKa = 3.64YY275 pKa = 10.1RR276 pKa = 11.84VRR278 pKa = 11.84FDD280 pKa = 3.99LDD282 pKa = 3.22NVAVASHH289 pKa = 5.21THH291 pKa = 5.52YY292 pKa = 10.63PVAKK296 pKa = 9.08DD297 pKa = 3.52TTWDD301 pKa = 3.69KK302 pKa = 9.29LTHH305 pKa = 5.48MASMLGNGVQDD316 pKa = 3.07IADD319 pKa = 3.41IVSTVGGAARR329 pKa = 11.84AMRR332 pKa = 11.84PYY334 pKa = 9.98ISSGSSLPMIAAA346 pKa = 4.56
MM1 pKa = 7.48AQRR4 pKa = 11.84KK5 pKa = 7.12SRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84NGNSQQNGQQNGQQLALRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84NGRR33 pKa = 11.84RR34 pKa = 11.84GPNARR39 pKa = 11.84TLVPGVGSTPGSPFGGIVGSDD60 pKa = 3.35LRR62 pKa = 11.84CWDD65 pKa = 3.46AKK67 pKa = 11.05LPLHH71 pKa = 6.59LPLPRR76 pKa = 11.84AVGPYY81 pKa = 9.14TVIRR85 pKa = 11.84CTRR88 pKa = 11.84RR89 pKa = 11.84INTANRR95 pKa = 11.84QMLFGTFQDD104 pKa = 4.08DD105 pKa = 3.99DD106 pKa = 4.08GTLGLVWNNICALGAINHH124 pKa = 6.83HH125 pKa = 6.85DD126 pKa = 4.91PINAANNTQRR136 pKa = 11.84YY137 pKa = 6.91TIDD140 pKa = 3.28LGGLGSGCSLVPAAMTVQVMNPNALQTTSGVIYY173 pKa = 10.15AGVMNTQAAFGGDD186 pKa = 3.03STAYY190 pKa = 10.26GDD192 pKa = 3.93RR193 pKa = 11.84FDD195 pKa = 4.76TFVEE199 pKa = 4.71FQNPRR204 pKa = 11.84LCAAPKK210 pKa = 9.52LALRR214 pKa = 11.84GVQMSSYY221 pKa = 8.83PLNMSKK227 pKa = 10.41VSEE230 pKa = 4.17FTGLGVATDD239 pKa = 4.75SVITWSDD246 pKa = 3.04PTLRR250 pKa = 11.84PTGWAPIMIYY260 pKa = 8.89NTAGTTLEE268 pKa = 3.94YY269 pKa = 10.61LVTIEE274 pKa = 3.64YY275 pKa = 10.1RR276 pKa = 11.84VRR278 pKa = 11.84FDD280 pKa = 3.99LDD282 pKa = 3.22NVAVASHH289 pKa = 5.21THH291 pKa = 5.52YY292 pKa = 10.63PVAKK296 pKa = 9.08DD297 pKa = 3.52TTWDD301 pKa = 3.69KK302 pKa = 9.29LTHH305 pKa = 5.48MASMLGNGVQDD316 pKa = 3.07IADD319 pKa = 3.41IVSTVGGAARR329 pKa = 11.84AMRR332 pKa = 11.84PYY334 pKa = 9.98ISSGSSLPMIAAA346 pKa = 4.56
Molecular weight: 37.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1161 |
346 |
815 |
580.5 |
64.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.527 ± 0.502 | 2.067 ± 0.309 |
6.546 ± 0.668 | 4.307 ± 1.566 |
3.704 ± 0.548 | 7.063 ± 1.517 |
2.67 ± 0.465 | 4.565 ± 0.029 |
3.359 ± 0.808 | 8.613 ± 0.259 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.101 ± 0.183 | 4.048 ± 1.148 |
5.857 ± 0.038 | 3.704 ± 0.457 |
7.666 ± 0.075 | 6.288 ± 0.035 |
6.632 ± 0.869 | 7.235 ± 0.148 |
1.637 ± 0.095 | 2.412 ± 0.238 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
