
Odonata-associated circular virus 21
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Bovismacovirus; Dragonfly associated bovismacovirus 1
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
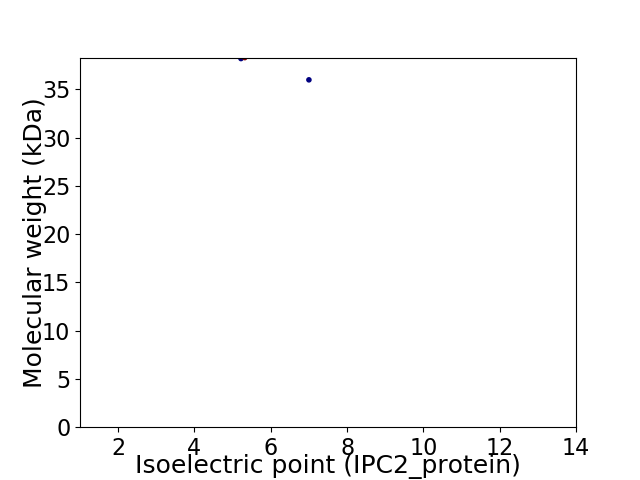
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UH63|A0A0B4UH63_9VIRU Putative capsid protein OS=Odonata-associated circular virus 21 OX=1592122 PE=4 SV=1
MM1 pKa = 7.79EE2 pKa = 6.22DD3 pKa = 3.32IIGAGLIGAGGNMVIVSVSEE23 pKa = 4.22TYY25 pKa = 10.78DD26 pKa = 4.29LSTKK30 pKa = 8.62PNKK33 pKa = 9.36MSLIALHH40 pKa = 5.93TPSRR44 pKa = 11.84EE45 pKa = 4.22IIKK48 pKa = 8.8KK49 pKa = 8.08TYY51 pKa = 9.93PGLCMNCKK59 pKa = 9.13YY60 pKa = 10.94VRR62 pKa = 11.84IVKK65 pKa = 9.96QDD67 pKa = 3.09VTMACASMLPADD79 pKa = 4.36PLQVGVSSGDD89 pKa = 3.31IAPQDD94 pKa = 3.6MFNPILYY101 pKa = 9.69KK102 pKa = 10.65AISTEE107 pKa = 3.97SLSILEE113 pKa = 3.96ARR115 pKa = 11.84IVGLQAQPSQNVFNGEE131 pKa = 4.37SVEE134 pKa = 4.27KK135 pKa = 10.31TDD137 pKa = 5.45DD138 pKa = 3.7ATNMADD144 pKa = 4.72DD145 pKa = 3.91FAVYY149 pKa = 10.15YY150 pKa = 10.91ALLSNRR156 pKa = 11.84DD157 pKa = 3.48GFRR160 pKa = 11.84TASPQAGFSMKK171 pKa = 10.54GLVPLVFEE179 pKa = 4.72KK180 pKa = 10.92YY181 pKa = 10.09YY182 pKa = 10.28PVGANSSSDD191 pKa = 3.07GNTAIIRR198 pKa = 11.84EE199 pKa = 3.99NSAGNGLEE207 pKa = 3.71VSTWSGRR214 pKa = 11.84SMRR217 pKa = 11.84GRR219 pKa = 11.84AHH221 pKa = 6.98PMPRR225 pKa = 11.84FNTTYY230 pKa = 9.04VTGVSKK236 pKa = 10.98AGEE239 pKa = 4.16TGVNRR244 pKa = 11.84QEE246 pKa = 4.17NGMTNGHH253 pKa = 7.36PYY255 pKa = 9.38NCQVEE260 pKa = 4.76MPTIPIIYY268 pKa = 8.61TGAIIMPPSKK278 pKa = 10.52LNQLYY283 pKa = 9.24YY284 pKa = 10.85RR285 pKa = 11.84MVVRR289 pKa = 11.84TYY291 pKa = 10.54IEE293 pKa = 3.99FSEE296 pKa = 4.1VRR298 pKa = 11.84PIQEE302 pKa = 3.3ISSFAEE308 pKa = 3.54LATYY312 pKa = 7.37YY313 pKa = 10.2TPEE316 pKa = 4.26VYY318 pKa = 10.53GSDD321 pKa = 3.42YY322 pKa = 10.68STVAKK327 pKa = 10.25TMTSQMAMVDD337 pKa = 3.68TQNADD342 pKa = 2.52ITKK345 pKa = 10.22IMDD348 pKa = 3.33GMM350 pKa = 4.22
MM1 pKa = 7.79EE2 pKa = 6.22DD3 pKa = 3.32IIGAGLIGAGGNMVIVSVSEE23 pKa = 4.22TYY25 pKa = 10.78DD26 pKa = 4.29LSTKK30 pKa = 8.62PNKK33 pKa = 9.36MSLIALHH40 pKa = 5.93TPSRR44 pKa = 11.84EE45 pKa = 4.22IIKK48 pKa = 8.8KK49 pKa = 8.08TYY51 pKa = 9.93PGLCMNCKK59 pKa = 9.13YY60 pKa = 10.94VRR62 pKa = 11.84IVKK65 pKa = 9.96QDD67 pKa = 3.09VTMACASMLPADD79 pKa = 4.36PLQVGVSSGDD89 pKa = 3.31IAPQDD94 pKa = 3.6MFNPILYY101 pKa = 9.69KK102 pKa = 10.65AISTEE107 pKa = 3.97SLSILEE113 pKa = 3.96ARR115 pKa = 11.84IVGLQAQPSQNVFNGEE131 pKa = 4.37SVEE134 pKa = 4.27KK135 pKa = 10.31TDD137 pKa = 5.45DD138 pKa = 3.7ATNMADD144 pKa = 4.72DD145 pKa = 3.91FAVYY149 pKa = 10.15YY150 pKa = 10.91ALLSNRR156 pKa = 11.84DD157 pKa = 3.48GFRR160 pKa = 11.84TASPQAGFSMKK171 pKa = 10.54GLVPLVFEE179 pKa = 4.72KK180 pKa = 10.92YY181 pKa = 10.09YY182 pKa = 10.28PVGANSSSDD191 pKa = 3.07GNTAIIRR198 pKa = 11.84EE199 pKa = 3.99NSAGNGLEE207 pKa = 3.71VSTWSGRR214 pKa = 11.84SMRR217 pKa = 11.84GRR219 pKa = 11.84AHH221 pKa = 6.98PMPRR225 pKa = 11.84FNTTYY230 pKa = 9.04VTGVSKK236 pKa = 10.98AGEE239 pKa = 4.16TGVNRR244 pKa = 11.84QEE246 pKa = 4.17NGMTNGHH253 pKa = 7.36PYY255 pKa = 9.38NCQVEE260 pKa = 4.76MPTIPIIYY268 pKa = 8.61TGAIIMPPSKK278 pKa = 10.52LNQLYY283 pKa = 9.24YY284 pKa = 10.85RR285 pKa = 11.84MVVRR289 pKa = 11.84TYY291 pKa = 10.54IEE293 pKa = 3.99FSEE296 pKa = 4.1VRR298 pKa = 11.84PIQEE302 pKa = 3.3ISSFAEE308 pKa = 3.54LATYY312 pKa = 7.37YY313 pKa = 10.2TPEE316 pKa = 4.26VYY318 pKa = 10.53GSDD321 pKa = 3.42YY322 pKa = 10.68STVAKK327 pKa = 10.25TMTSQMAMVDD337 pKa = 3.68TQNADD342 pKa = 2.52ITKK345 pKa = 10.22IMDD348 pKa = 3.33GMM350 pKa = 4.22
Molecular weight: 38.18 kDa
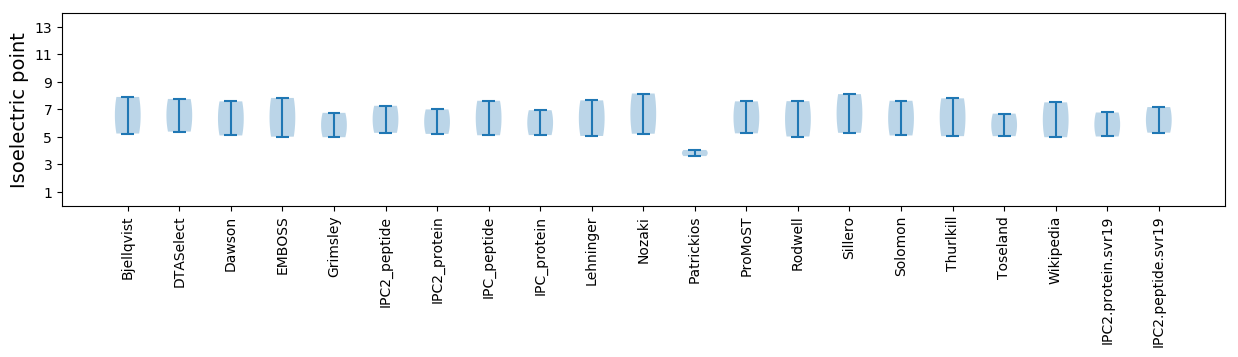
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UH63|A0A0B4UH63_9VIRU Putative capsid protein OS=Odonata-associated circular virus 21 OX=1592122 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 10.17KK3 pKa = 10.41CEE5 pKa = 4.01ACEE8 pKa = 3.86EE9 pKa = 4.19RR10 pKa = 11.84PANYY14 pKa = 10.34GYY16 pKa = 10.78VKK18 pKa = 10.09WINCIDD24 pKa = 4.24GIDD27 pKa = 4.05RR28 pKa = 11.84YY29 pKa = 9.67WFGYY33 pKa = 9.78ICDD36 pKa = 3.59EE37 pKa = 4.7CYY39 pKa = 10.66HH40 pKa = 6.86KK41 pKa = 11.02FEE43 pKa = 4.64EE44 pKa = 4.61EE45 pKa = 3.65WAKK48 pKa = 10.96EE49 pKa = 4.01EE50 pKa = 4.15EE51 pKa = 4.85CNVQYY56 pKa = 10.66IGTISAEE63 pKa = 4.07DD64 pKa = 3.47WNEE67 pKa = 3.59DD68 pKa = 3.31GIIRR72 pKa = 11.84VFEE75 pKa = 4.2NLDD78 pKa = 3.27GHH80 pKa = 5.97EE81 pKa = 4.38LYY83 pKa = 10.25IGRR86 pKa = 11.84EE87 pKa = 3.76IGKK90 pKa = 9.23HH91 pKa = 4.58GFRR94 pKa = 11.84HH95 pKa = 5.35YY96 pKa = 10.14QFCMDD101 pKa = 4.44CAGDD105 pKa = 3.94LEE107 pKa = 6.03KK108 pKa = 10.77YY109 pKa = 8.37TADD112 pKa = 4.29NRR114 pKa = 11.84TGWHH118 pKa = 5.16VEE120 pKa = 3.95RR121 pKa = 11.84CISWEE126 pKa = 3.82MSKK129 pKa = 10.78GYY131 pKa = 9.57CRR133 pKa = 11.84KK134 pKa = 8.5TGDD137 pKa = 3.11YY138 pKa = 10.34RR139 pKa = 11.84YY140 pKa = 9.98IGDD143 pKa = 3.75SRR145 pKa = 11.84EE146 pKa = 3.58EE147 pKa = 4.21RR148 pKa = 11.84YY149 pKa = 9.81YY150 pKa = 10.56AWLRR154 pKa = 11.84ARR156 pKa = 11.84RR157 pKa = 11.84PLPIWRR163 pKa = 11.84SFGASVVKK171 pKa = 10.89QNDD174 pKa = 2.95RR175 pKa = 11.84SITVWVDD182 pKa = 2.99TEE184 pKa = 4.33GKK186 pKa = 9.93AGKK189 pKa = 8.29STFSYY194 pKa = 10.12ILEE197 pKa = 4.16RR198 pKa = 11.84RR199 pKa = 11.84GKK201 pKa = 9.78CLNIPRR207 pKa = 11.84TEE209 pKa = 4.15HH210 pKa = 6.04NPTRR214 pKa = 11.84LIDD217 pKa = 3.91FVAMHH222 pKa = 6.19YY223 pKa = 10.22KK224 pKa = 10.51GEE226 pKa = 4.02PLIIVDD232 pKa = 4.16IPRR235 pKa = 11.84DD236 pKa = 3.51QKK238 pKa = 11.07LGKK241 pKa = 8.52EE242 pKa = 4.06LCRR245 pKa = 11.84ALEE248 pKa = 4.28TIKK251 pKa = 11.17DD252 pKa = 3.92GVITSAKK259 pKa = 8.61YY260 pKa = 9.95QGTKK264 pKa = 9.38MFIKK268 pKa = 10.25GVKK271 pKa = 9.37ILVFTNHH278 pKa = 6.3FLDD281 pKa = 3.48KK282 pKa = 8.73TTYY285 pKa = 10.68AALTEE290 pKa = 4.39DD291 pKa = 3.13RR292 pKa = 11.84WDD294 pKa = 3.74VKK296 pKa = 9.99SLKK299 pKa = 10.11ASKK302 pKa = 9.97SHH304 pKa = 6.41PRR306 pKa = 3.28
MM1 pKa = 7.45KK2 pKa = 10.17KK3 pKa = 10.41CEE5 pKa = 4.01ACEE8 pKa = 3.86EE9 pKa = 4.19RR10 pKa = 11.84PANYY14 pKa = 10.34GYY16 pKa = 10.78VKK18 pKa = 10.09WINCIDD24 pKa = 4.24GIDD27 pKa = 4.05RR28 pKa = 11.84YY29 pKa = 9.67WFGYY33 pKa = 9.78ICDD36 pKa = 3.59EE37 pKa = 4.7CYY39 pKa = 10.66HH40 pKa = 6.86KK41 pKa = 11.02FEE43 pKa = 4.64EE44 pKa = 4.61EE45 pKa = 3.65WAKK48 pKa = 10.96EE49 pKa = 4.01EE50 pKa = 4.15EE51 pKa = 4.85CNVQYY56 pKa = 10.66IGTISAEE63 pKa = 4.07DD64 pKa = 3.47WNEE67 pKa = 3.59DD68 pKa = 3.31GIIRR72 pKa = 11.84VFEE75 pKa = 4.2NLDD78 pKa = 3.27GHH80 pKa = 5.97EE81 pKa = 4.38LYY83 pKa = 10.25IGRR86 pKa = 11.84EE87 pKa = 3.76IGKK90 pKa = 9.23HH91 pKa = 4.58GFRR94 pKa = 11.84HH95 pKa = 5.35YY96 pKa = 10.14QFCMDD101 pKa = 4.44CAGDD105 pKa = 3.94LEE107 pKa = 6.03KK108 pKa = 10.77YY109 pKa = 8.37TADD112 pKa = 4.29NRR114 pKa = 11.84TGWHH118 pKa = 5.16VEE120 pKa = 3.95RR121 pKa = 11.84CISWEE126 pKa = 3.82MSKK129 pKa = 10.78GYY131 pKa = 9.57CRR133 pKa = 11.84KK134 pKa = 8.5TGDD137 pKa = 3.11YY138 pKa = 10.34RR139 pKa = 11.84YY140 pKa = 9.98IGDD143 pKa = 3.75SRR145 pKa = 11.84EE146 pKa = 3.58EE147 pKa = 4.21RR148 pKa = 11.84YY149 pKa = 9.81YY150 pKa = 10.56AWLRR154 pKa = 11.84ARR156 pKa = 11.84RR157 pKa = 11.84PLPIWRR163 pKa = 11.84SFGASVVKK171 pKa = 10.89QNDD174 pKa = 2.95RR175 pKa = 11.84SITVWVDD182 pKa = 2.99TEE184 pKa = 4.33GKK186 pKa = 9.93AGKK189 pKa = 8.29STFSYY194 pKa = 10.12ILEE197 pKa = 4.16RR198 pKa = 11.84RR199 pKa = 11.84GKK201 pKa = 9.78CLNIPRR207 pKa = 11.84TEE209 pKa = 4.15HH210 pKa = 6.04NPTRR214 pKa = 11.84LIDD217 pKa = 3.91FVAMHH222 pKa = 6.19YY223 pKa = 10.22KK224 pKa = 10.51GEE226 pKa = 4.02PLIIVDD232 pKa = 4.16IPRR235 pKa = 11.84DD236 pKa = 3.51QKK238 pKa = 11.07LGKK241 pKa = 8.52EE242 pKa = 4.06LCRR245 pKa = 11.84ALEE248 pKa = 4.28TIKK251 pKa = 11.17DD252 pKa = 3.92GVITSAKK259 pKa = 8.61YY260 pKa = 9.95QGTKK264 pKa = 9.38MFIKK268 pKa = 10.25GVKK271 pKa = 9.37ILVFTNHH278 pKa = 6.3FLDD281 pKa = 3.48KK282 pKa = 8.73TTYY285 pKa = 10.68AALTEE290 pKa = 4.39DD291 pKa = 3.13RR292 pKa = 11.84WDD294 pKa = 3.74VKK296 pKa = 9.99SLKK299 pKa = 10.11ASKK302 pKa = 9.97SHH304 pKa = 6.41PRR306 pKa = 3.28
Molecular weight: 35.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
656 |
306 |
350 |
328.0 |
37.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.555 ± 0.913 | 2.439 ± 1.02 |
5.488 ± 0.721 | 6.86 ± 1.352 |
3.049 ± 0.376 | 7.622 ± 0.073 |
1.829 ± 0.765 | 7.47 ± 0.257 |
5.945 ± 1.531 | 5.335 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.811 ± 1.498 | 4.421 ± 0.793 |
4.268 ± 1.138 | 2.744 ± 0.764 |
5.793 ± 1.411 | 6.707 ± 1.693 |
6.402 ± 0.808 | 6.098 ± 1.048 |
1.677 ± 1.095 | 5.488 ± 0.272 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
