
Lake Sinai virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Magsaviricetes; Nodamuvirales; Sinhaliviridae; Sinaivirus; unclassified Sinaivirus
Average proteome isoelectric point is 7.72
Get precalculated fractions of proteins
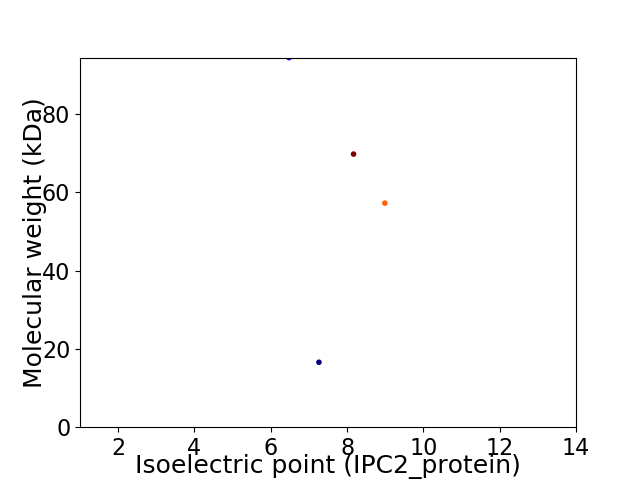
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGU1|A0A1L3KGU1_9VIRU Chropara_Vmeth domain-containing protein OS=Lake Sinai virus OX=1547219 PE=4 SV=1
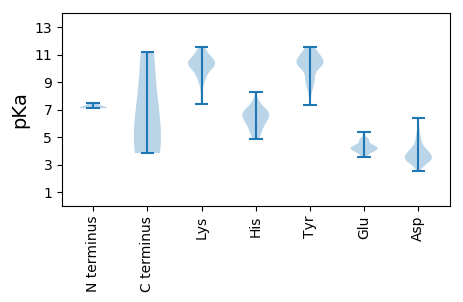
MM1 pKa = 7.12VKK3 pKa = 10.35LVFAAFIALYY13 pKa = 9.16LVCQVPPISHH23 pKa = 7.43WIEE26 pKa = 3.68DD27 pKa = 4.15WIDD30 pKa = 4.63DD31 pKa = 4.07FDD33 pKa = 4.31TMCSYY38 pKa = 10.99EE39 pKa = 4.2YY40 pKa = 10.97AAADD44 pKa = 3.72AYY46 pKa = 11.49NSFVHH51 pKa = 5.45KK52 pKa = 10.37HH53 pKa = 4.85RR54 pKa = 11.84VAAYY58 pKa = 7.84AAGVRR63 pKa = 11.84VYY65 pKa = 10.38RR66 pKa = 11.84YY67 pKa = 7.31RR68 pKa = 11.84TPWYY72 pKa = 8.93CVASRR77 pKa = 11.84TVPVVHH83 pKa = 6.67PFGWLMAKK91 pKa = 10.29YY92 pKa = 10.03EE93 pKa = 4.29STTYY97 pKa = 10.28HH98 pKa = 6.0VSEE101 pKa = 4.42LLDD104 pKa = 3.4RR105 pKa = 11.84LGIANATLSDD115 pKa = 3.58GLRR118 pKa = 11.84AVAEE122 pKa = 4.38TAFTAYY128 pKa = 9.11MYY130 pKa = 10.17YY131 pKa = 10.15QIVWFQLVFIILLTGSFIAIVCSSRR156 pKa = 11.84VRR158 pKa = 11.84VVRR161 pKa = 11.84VRR163 pKa = 11.84NRR165 pKa = 11.84VRR167 pKa = 11.84GYY169 pKa = 10.6SVAEE173 pKa = 3.74LRR175 pKa = 11.84AEE177 pKa = 4.06FEE179 pKa = 4.43DD180 pKa = 5.54AISDD184 pKa = 3.87VNAPPSRR191 pKa = 11.84GHH193 pKa = 6.65SGLTFQRR200 pKa = 11.84KK201 pKa = 7.41VVEE204 pKa = 4.48SWCLDD209 pKa = 3.05QLLRR213 pKa = 11.84YY214 pKa = 8.25FRR216 pKa = 11.84HH217 pKa = 5.0VRR219 pKa = 11.84FVSATCDD226 pKa = 2.59RR227 pKa = 11.84WAEE230 pKa = 3.93LGHH233 pKa = 6.76RR234 pKa = 11.84IHH236 pKa = 7.02RR237 pKa = 11.84CAPVVMDD244 pKa = 4.08GSFVPEE250 pKa = 4.03THH252 pKa = 6.69HH253 pKa = 6.98GGVGCRR259 pKa = 11.84RR260 pKa = 11.84APSNCPDD267 pKa = 3.78RR268 pKa = 11.84YY269 pKa = 10.66DD270 pKa = 3.56IPACVLSHH278 pKa = 6.23VDD280 pKa = 3.79YY281 pKa = 11.58YY282 pKa = 10.93MSPDD286 pKa = 3.42QLASTVVGPTFIVNHH301 pKa = 7.23DD302 pKa = 3.91YY303 pKa = 11.57SCVDD307 pKa = 3.4SLSVAEE313 pKa = 4.2ANIRR317 pKa = 11.84AAGGLITSSVRR328 pKa = 11.84DD329 pKa = 3.74GPVYY333 pKa = 10.58GPHH336 pKa = 7.68PYY338 pKa = 10.22HH339 pKa = 6.59LWDD342 pKa = 3.65NEE344 pKa = 4.13GVVVSSTGAFRR355 pKa = 11.84YY356 pKa = 9.71YY357 pKa = 10.35RR358 pKa = 11.84VGRR361 pKa = 11.84LYY363 pKa = 10.46DD364 pKa = 3.3TTLYY368 pKa = 10.9YY369 pKa = 10.9AFPVAGTYY377 pKa = 9.23TRR379 pKa = 11.84DD380 pKa = 3.27DD381 pKa = 3.88PTNLRR386 pKa = 11.84CSTVGDD392 pKa = 3.2FHH394 pKa = 8.25YY395 pKa = 10.72YY396 pKa = 9.93SPHH399 pKa = 5.6EE400 pKa = 4.2KK401 pKa = 10.45RR402 pKa = 11.84FISYY406 pKa = 10.41SVDD409 pKa = 2.91GNSYY413 pKa = 9.94HH414 pKa = 6.11VFGVSVPRR422 pKa = 11.84SLADD426 pKa = 3.09YY427 pKa = 10.3CAATFCRR434 pKa = 11.84SARR437 pKa = 11.84DD438 pKa = 3.55DD439 pKa = 3.42KK440 pKa = 11.38FYY442 pKa = 11.33DD443 pKa = 3.57SLRR446 pKa = 11.84SYY448 pKa = 9.17YY449 pKa = 9.02QNRR452 pKa = 11.84CRR454 pKa = 11.84AIGFTDD460 pKa = 3.92ARR462 pKa = 11.84DD463 pKa = 3.62TLILDD468 pKa = 4.81FIIHH472 pKa = 6.66LCDD475 pKa = 3.16EE476 pKa = 4.87ASLKK480 pKa = 10.08TFGFSRR486 pKa = 11.84LSTAPSSWTAYY497 pKa = 8.96CLSWVLVKK505 pKa = 10.45FNHH508 pKa = 6.01IMPLALTSFTVNALHH523 pKa = 7.11RR524 pKa = 11.84FFGAKK529 pKa = 8.97AAPWNWASIHH539 pKa = 6.35LPTYY543 pKa = 11.51DD544 pKa = 3.08MVTSPFRR551 pKa = 11.84LKK553 pKa = 10.38MFGRR557 pKa = 11.84NPTTFNLEE565 pKa = 3.83RR566 pKa = 11.84FRR568 pKa = 11.84AEE570 pKa = 3.81ATVVGAPNLGQSAAGASQDD589 pKa = 3.75SDD591 pKa = 3.19KK592 pKa = 11.55HH593 pKa = 6.64DD594 pKa = 3.47IQLGNPGAAAGSVSSSTFSDD614 pKa = 3.49SASASDD620 pKa = 5.72DD621 pKa = 3.33VLLDD625 pKa = 3.88HH626 pKa = 7.15EE627 pKa = 5.36SGSILDD633 pKa = 5.05DD634 pKa = 4.07SFCSSSPTYY643 pKa = 10.41GSSGVLSNKK652 pKa = 9.35GKK654 pKa = 10.45ARR656 pKa = 11.84RR657 pKa = 11.84KK658 pKa = 7.82PHH660 pKa = 6.73RR661 pKa = 11.84PTNRR665 pKa = 11.84DD666 pKa = 2.98PDD668 pKa = 3.62RR669 pKa = 11.84DD670 pKa = 3.64QEE672 pKa = 4.79HH673 pKa = 6.94NICHH677 pKa = 6.99PKK679 pKa = 8.42RR680 pKa = 11.84TTYY683 pKa = 11.18ADD685 pKa = 3.91CPDD688 pKa = 3.66PTASCGPHH696 pKa = 6.48FFMSVCEE703 pKa = 4.06RR704 pKa = 11.84DD705 pKa = 3.33EE706 pKa = 4.23SVPTLFHH713 pKa = 5.9AHH715 pKa = 6.13AVGGEE720 pKa = 4.36DD721 pKa = 2.69ITHH724 pKa = 5.31VVEE727 pKa = 4.84PDD729 pKa = 2.96LGIAISEE736 pKa = 4.25RR737 pKa = 11.84FSASQLRR744 pKa = 11.84LLSWSIDD751 pKa = 3.36GVFNTLSRR759 pKa = 11.84AATSSFVEE767 pKa = 4.23SSLLSLLRR775 pKa = 11.84FMQQVPTTQPSAEE788 pKa = 3.96ARR790 pKa = 11.84RR791 pKa = 11.84CLLFGGPRR799 pKa = 11.84QRR801 pKa = 11.84FKK803 pKa = 11.27GYY805 pKa = 8.63DD806 pKa = 3.27TFDD809 pKa = 3.68LGFMGISVPTSKK821 pKa = 9.56TAGVASCLRR830 pKa = 11.84EE831 pKa = 3.94VARR834 pKa = 11.84QHH836 pKa = 6.28ACADD840 pKa = 3.94SPHH843 pKa = 6.91EE844 pKa = 4.38GPKK847 pKa = 10.29LHH849 pKa = 6.95
MM1 pKa = 7.12VKK3 pKa = 10.35LVFAAFIALYY13 pKa = 9.16LVCQVPPISHH23 pKa = 7.43WIEE26 pKa = 3.68DD27 pKa = 4.15WIDD30 pKa = 4.63DD31 pKa = 4.07FDD33 pKa = 4.31TMCSYY38 pKa = 10.99EE39 pKa = 4.2YY40 pKa = 10.97AAADD44 pKa = 3.72AYY46 pKa = 11.49NSFVHH51 pKa = 5.45KK52 pKa = 10.37HH53 pKa = 4.85RR54 pKa = 11.84VAAYY58 pKa = 7.84AAGVRR63 pKa = 11.84VYY65 pKa = 10.38RR66 pKa = 11.84YY67 pKa = 7.31RR68 pKa = 11.84TPWYY72 pKa = 8.93CVASRR77 pKa = 11.84TVPVVHH83 pKa = 6.67PFGWLMAKK91 pKa = 10.29YY92 pKa = 10.03EE93 pKa = 4.29STTYY97 pKa = 10.28HH98 pKa = 6.0VSEE101 pKa = 4.42LLDD104 pKa = 3.4RR105 pKa = 11.84LGIANATLSDD115 pKa = 3.58GLRR118 pKa = 11.84AVAEE122 pKa = 4.38TAFTAYY128 pKa = 9.11MYY130 pKa = 10.17YY131 pKa = 10.15QIVWFQLVFIILLTGSFIAIVCSSRR156 pKa = 11.84VRR158 pKa = 11.84VVRR161 pKa = 11.84VRR163 pKa = 11.84NRR165 pKa = 11.84VRR167 pKa = 11.84GYY169 pKa = 10.6SVAEE173 pKa = 3.74LRR175 pKa = 11.84AEE177 pKa = 4.06FEE179 pKa = 4.43DD180 pKa = 5.54AISDD184 pKa = 3.87VNAPPSRR191 pKa = 11.84GHH193 pKa = 6.65SGLTFQRR200 pKa = 11.84KK201 pKa = 7.41VVEE204 pKa = 4.48SWCLDD209 pKa = 3.05QLLRR213 pKa = 11.84YY214 pKa = 8.25FRR216 pKa = 11.84HH217 pKa = 5.0VRR219 pKa = 11.84FVSATCDD226 pKa = 2.59RR227 pKa = 11.84WAEE230 pKa = 3.93LGHH233 pKa = 6.76RR234 pKa = 11.84IHH236 pKa = 7.02RR237 pKa = 11.84CAPVVMDD244 pKa = 4.08GSFVPEE250 pKa = 4.03THH252 pKa = 6.69HH253 pKa = 6.98GGVGCRR259 pKa = 11.84RR260 pKa = 11.84APSNCPDD267 pKa = 3.78RR268 pKa = 11.84YY269 pKa = 10.66DD270 pKa = 3.56IPACVLSHH278 pKa = 6.23VDD280 pKa = 3.79YY281 pKa = 11.58YY282 pKa = 10.93MSPDD286 pKa = 3.42QLASTVVGPTFIVNHH301 pKa = 7.23DD302 pKa = 3.91YY303 pKa = 11.57SCVDD307 pKa = 3.4SLSVAEE313 pKa = 4.2ANIRR317 pKa = 11.84AAGGLITSSVRR328 pKa = 11.84DD329 pKa = 3.74GPVYY333 pKa = 10.58GPHH336 pKa = 7.68PYY338 pKa = 10.22HH339 pKa = 6.59LWDD342 pKa = 3.65NEE344 pKa = 4.13GVVVSSTGAFRR355 pKa = 11.84YY356 pKa = 9.71YY357 pKa = 10.35RR358 pKa = 11.84VGRR361 pKa = 11.84LYY363 pKa = 10.46DD364 pKa = 3.3TTLYY368 pKa = 10.9YY369 pKa = 10.9AFPVAGTYY377 pKa = 9.23TRR379 pKa = 11.84DD380 pKa = 3.27DD381 pKa = 3.88PTNLRR386 pKa = 11.84CSTVGDD392 pKa = 3.2FHH394 pKa = 8.25YY395 pKa = 10.72YY396 pKa = 9.93SPHH399 pKa = 5.6EE400 pKa = 4.2KK401 pKa = 10.45RR402 pKa = 11.84FISYY406 pKa = 10.41SVDD409 pKa = 2.91GNSYY413 pKa = 9.94HH414 pKa = 6.11VFGVSVPRR422 pKa = 11.84SLADD426 pKa = 3.09YY427 pKa = 10.3CAATFCRR434 pKa = 11.84SARR437 pKa = 11.84DD438 pKa = 3.55DD439 pKa = 3.42KK440 pKa = 11.38FYY442 pKa = 11.33DD443 pKa = 3.57SLRR446 pKa = 11.84SYY448 pKa = 9.17YY449 pKa = 9.02QNRR452 pKa = 11.84CRR454 pKa = 11.84AIGFTDD460 pKa = 3.92ARR462 pKa = 11.84DD463 pKa = 3.62TLILDD468 pKa = 4.81FIIHH472 pKa = 6.66LCDD475 pKa = 3.16EE476 pKa = 4.87ASLKK480 pKa = 10.08TFGFSRR486 pKa = 11.84LSTAPSSWTAYY497 pKa = 8.96CLSWVLVKK505 pKa = 10.45FNHH508 pKa = 6.01IMPLALTSFTVNALHH523 pKa = 7.11RR524 pKa = 11.84FFGAKK529 pKa = 8.97AAPWNWASIHH539 pKa = 6.35LPTYY543 pKa = 11.51DD544 pKa = 3.08MVTSPFRR551 pKa = 11.84LKK553 pKa = 10.38MFGRR557 pKa = 11.84NPTTFNLEE565 pKa = 3.83RR566 pKa = 11.84FRR568 pKa = 11.84AEE570 pKa = 3.81ATVVGAPNLGQSAAGASQDD589 pKa = 3.75SDD591 pKa = 3.19KK592 pKa = 11.55HH593 pKa = 6.64DD594 pKa = 3.47IQLGNPGAAAGSVSSSTFSDD614 pKa = 3.49SASASDD620 pKa = 5.72DD621 pKa = 3.33VLLDD625 pKa = 3.88HH626 pKa = 7.15EE627 pKa = 5.36SGSILDD633 pKa = 5.05DD634 pKa = 4.07SFCSSSPTYY643 pKa = 10.41GSSGVLSNKK652 pKa = 9.35GKK654 pKa = 10.45ARR656 pKa = 11.84RR657 pKa = 11.84KK658 pKa = 7.82PHH660 pKa = 6.73RR661 pKa = 11.84PTNRR665 pKa = 11.84DD666 pKa = 2.98PDD668 pKa = 3.62RR669 pKa = 11.84DD670 pKa = 3.64QEE672 pKa = 4.79HH673 pKa = 6.94NICHH677 pKa = 6.99PKK679 pKa = 8.42RR680 pKa = 11.84TTYY683 pKa = 11.18ADD685 pKa = 3.91CPDD688 pKa = 3.66PTASCGPHH696 pKa = 6.48FFMSVCEE703 pKa = 4.06RR704 pKa = 11.84DD705 pKa = 3.33EE706 pKa = 4.23SVPTLFHH713 pKa = 5.9AHH715 pKa = 6.13AVGGEE720 pKa = 4.36DD721 pKa = 2.69ITHH724 pKa = 5.31VVEE727 pKa = 4.84PDD729 pKa = 2.96LGIAISEE736 pKa = 4.25RR737 pKa = 11.84FSASQLRR744 pKa = 11.84LLSWSIDD751 pKa = 3.36GVFNTLSRR759 pKa = 11.84AATSSFVEE767 pKa = 4.23SSLLSLLRR775 pKa = 11.84FMQQVPTTQPSAEE788 pKa = 3.96ARR790 pKa = 11.84RR791 pKa = 11.84CLLFGGPRR799 pKa = 11.84QRR801 pKa = 11.84FKK803 pKa = 11.27GYY805 pKa = 8.63DD806 pKa = 3.27TFDD809 pKa = 3.68LGFMGISVPTSKK821 pKa = 9.56TAGVASCLRR830 pKa = 11.84EE831 pKa = 3.94VARR834 pKa = 11.84QHH836 pKa = 6.28ACADD840 pKa = 3.94SPHH843 pKa = 6.91EE844 pKa = 4.38GPKK847 pKa = 10.29LHH849 pKa = 6.95
Molecular weight: 94.48 kDa
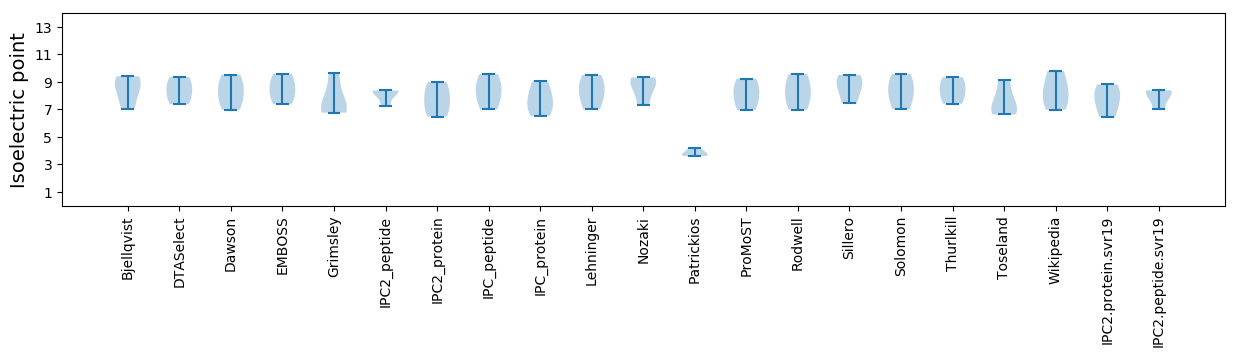
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGN5|A0A1L3KGN5_9VIRU RNA-directed RNA polymerase OS=Lake Sinai virus OX=1547219 PE=4 SV=1
MM1 pKa = 7.17NQQQSTASPRR11 pKa = 11.84LARR14 pKa = 11.84PSPQAPTSRR23 pKa = 11.84SARR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84NRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84NGATAAVGPVAIQPSRR50 pKa = 11.84ITRR53 pKa = 11.84RR54 pKa = 11.84TVTNLTRR61 pKa = 11.84RR62 pKa = 11.84PFAITSAGLAWLRR75 pKa = 11.84QYY77 pKa = 10.73LNPMGPSMSSVAGFPDD93 pKa = 4.59GSAVTTCIADD103 pKa = 3.28YY104 pKa = 11.2SNNFNISFPPRR115 pKa = 11.84EE116 pKa = 4.35AIYY119 pKa = 8.7CTGTNSEE126 pKa = 4.18NQATLLDD133 pKa = 3.72AATYY137 pKa = 10.82AKK139 pKa = 9.89IDD141 pKa = 3.2AWTKK145 pKa = 11.36ADD147 pKa = 3.2ITLCILALPMLRR159 pKa = 11.84NVAMVRR165 pKa = 11.84LYY167 pKa = 8.26PTTPTAFTLSDD178 pKa = 4.74GIPNFVQRR186 pKa = 11.84FPNWSAFTTEE196 pKa = 5.19GKK198 pKa = 10.24LLNNGDD204 pKa = 4.01SPGYY208 pKa = 8.33IQSFIYY214 pKa = 10.08LPNVDD219 pKa = 3.94KK220 pKa = 11.02HH221 pKa = 6.81LSAARR226 pKa = 11.84GYY228 pKa = 10.13RR229 pKa = 11.84LLSRR233 pKa = 11.84GLTGIFTAPSLEE245 pKa = 4.15TQGFVTACQYY255 pKa = 10.41LAQGTVLTQTVGNDD269 pKa = 3.17FVQSVEE275 pKa = 4.04VNADD279 pKa = 3.33KK280 pKa = 10.45TVKK283 pKa = 10.27NVAGKK288 pKa = 9.55RR289 pKa = 11.84LHH291 pKa = 6.32YY292 pKa = 10.3SGPPKK297 pKa = 10.55YY298 pKa = 10.45VFPLEE303 pKa = 4.48GDD305 pKa = 3.17NCAPSSLVEE314 pKa = 4.57TYY316 pKa = 10.02HH317 pKa = 6.01QAYY320 pKa = 8.56QARR323 pKa = 11.84AVDD326 pKa = 4.3GFYY329 pKa = 10.36MPLLSSSRR337 pKa = 11.84DD338 pKa = 3.45NPFQSPRR345 pKa = 11.84PQPIAVYY352 pKa = 10.15NRR354 pKa = 11.84WYY356 pKa = 10.9YY357 pKa = 10.44RR358 pKa = 11.84GCLDD362 pKa = 5.74PIPASKK368 pKa = 10.77YY369 pKa = 10.32ADD371 pKa = 3.73GPSQYY376 pKa = 10.58FYY378 pKa = 11.45DD379 pKa = 4.93LNLADD384 pKa = 5.86DD385 pKa = 4.13VAPLYY390 pKa = 9.08NTGVVWMEE398 pKa = 4.18GISPNFSLKK407 pKa = 10.69LKK409 pKa = 8.98TRR411 pKa = 11.84TVIQYY416 pKa = 9.27IPTSGSVLANFTRR429 pKa = 11.84HH430 pKa = 5.55EE431 pKa = 4.0PTYY434 pKa = 11.03DD435 pKa = 4.04QIALDD440 pKa = 3.51AADD443 pKa = 4.57RR444 pKa = 11.84VRR446 pKa = 11.84SMMPHH451 pKa = 7.71AYY453 pKa = 8.62PAAYY457 pKa = 9.97NDD459 pKa = 3.89WGWLGDD465 pKa = 4.28LLDD468 pKa = 4.4STLSTLPGISMAYY481 pKa = 9.97RR482 pKa = 11.84FAKK485 pKa = 10.2PLIKK489 pKa = 9.98PAWNWLGGKK498 pKa = 9.81VSDD501 pKa = 4.29FFGNPVSRR509 pKa = 11.84DD510 pKa = 2.54GDD512 pKa = 3.36IFYY515 pKa = 10.47DD516 pKa = 3.98AKK518 pKa = 11.17
MM1 pKa = 7.17NQQQSTASPRR11 pKa = 11.84LARR14 pKa = 11.84PSPQAPTSRR23 pKa = 11.84SARR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84NRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84NGATAAVGPVAIQPSRR50 pKa = 11.84ITRR53 pKa = 11.84RR54 pKa = 11.84TVTNLTRR61 pKa = 11.84RR62 pKa = 11.84PFAITSAGLAWLRR75 pKa = 11.84QYY77 pKa = 10.73LNPMGPSMSSVAGFPDD93 pKa = 4.59GSAVTTCIADD103 pKa = 3.28YY104 pKa = 11.2SNNFNISFPPRR115 pKa = 11.84EE116 pKa = 4.35AIYY119 pKa = 8.7CTGTNSEE126 pKa = 4.18NQATLLDD133 pKa = 3.72AATYY137 pKa = 10.82AKK139 pKa = 9.89IDD141 pKa = 3.2AWTKK145 pKa = 11.36ADD147 pKa = 3.2ITLCILALPMLRR159 pKa = 11.84NVAMVRR165 pKa = 11.84LYY167 pKa = 8.26PTTPTAFTLSDD178 pKa = 4.74GIPNFVQRR186 pKa = 11.84FPNWSAFTTEE196 pKa = 5.19GKK198 pKa = 10.24LLNNGDD204 pKa = 4.01SPGYY208 pKa = 8.33IQSFIYY214 pKa = 10.08LPNVDD219 pKa = 3.94KK220 pKa = 11.02HH221 pKa = 6.81LSAARR226 pKa = 11.84GYY228 pKa = 10.13RR229 pKa = 11.84LLSRR233 pKa = 11.84GLTGIFTAPSLEE245 pKa = 4.15TQGFVTACQYY255 pKa = 10.41LAQGTVLTQTVGNDD269 pKa = 3.17FVQSVEE275 pKa = 4.04VNADD279 pKa = 3.33KK280 pKa = 10.45TVKK283 pKa = 10.27NVAGKK288 pKa = 9.55RR289 pKa = 11.84LHH291 pKa = 6.32YY292 pKa = 10.3SGPPKK297 pKa = 10.55YY298 pKa = 10.45VFPLEE303 pKa = 4.48GDD305 pKa = 3.17NCAPSSLVEE314 pKa = 4.57TYY316 pKa = 10.02HH317 pKa = 6.01QAYY320 pKa = 8.56QARR323 pKa = 11.84AVDD326 pKa = 4.3GFYY329 pKa = 10.36MPLLSSSRR337 pKa = 11.84DD338 pKa = 3.45NPFQSPRR345 pKa = 11.84PQPIAVYY352 pKa = 10.15NRR354 pKa = 11.84WYY356 pKa = 10.9YY357 pKa = 10.44RR358 pKa = 11.84GCLDD362 pKa = 5.74PIPASKK368 pKa = 10.77YY369 pKa = 10.32ADD371 pKa = 3.73GPSQYY376 pKa = 10.58FYY378 pKa = 11.45DD379 pKa = 4.93LNLADD384 pKa = 5.86DD385 pKa = 4.13VAPLYY390 pKa = 9.08NTGVVWMEE398 pKa = 4.18GISPNFSLKK407 pKa = 10.69LKK409 pKa = 8.98TRR411 pKa = 11.84TVIQYY416 pKa = 9.27IPTSGSVLANFTRR429 pKa = 11.84HH430 pKa = 5.55EE431 pKa = 4.0PTYY434 pKa = 11.03DD435 pKa = 4.04QIALDD440 pKa = 3.51AADD443 pKa = 4.57RR444 pKa = 11.84VRR446 pKa = 11.84SMMPHH451 pKa = 7.71AYY453 pKa = 8.62PAAYY457 pKa = 9.97NDD459 pKa = 3.89WGWLGDD465 pKa = 4.28LLDD468 pKa = 4.4STLSTLPGISMAYY481 pKa = 9.97RR482 pKa = 11.84FAKK485 pKa = 10.2PLIKK489 pKa = 9.98PAWNWLGGKK498 pKa = 9.81VSDD501 pKa = 4.29FFGNPVSRR509 pKa = 11.84DD510 pKa = 2.54GDD512 pKa = 3.36IFYY515 pKa = 10.47DD516 pKa = 3.98AKK518 pKa = 11.17
Molecular weight: 57.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2139 |
150 |
849 |
534.8 |
59.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.883 ± 0.644 | 2.665 ± 0.454 |
5.563 ± 0.493 | 2.899 ± 0.386 |
4.348 ± 0.614 | 5.517 ± 0.469 |
2.805 ± 0.623 | 4.254 ± 0.391 |
2.478 ± 0.382 | 9.724 ± 1.558 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.823 ± 0.198 | 3.226 ± 0.716 |
6.498 ± 0.649 | 2.712 ± 0.462 |
7.387 ± 0.248 | 9.21 ± 0.476 |
6.498 ± 0.339 | 7.246 ± 0.639 |
1.449 ± 0.131 | 4.815 ± 0.14 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
