
Changjiang tombus-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.83
Get precalculated fractions of proteins
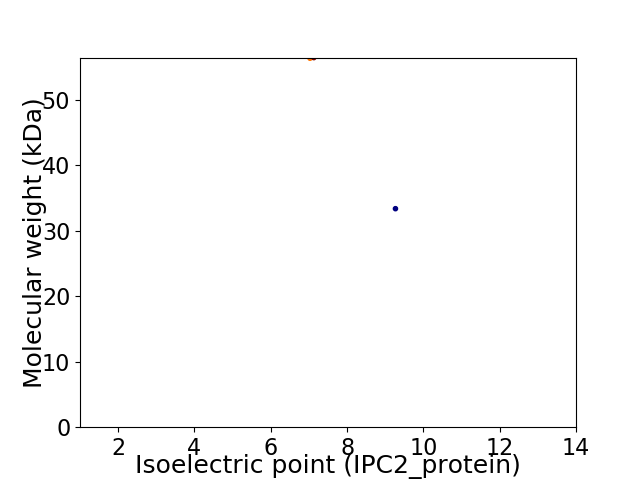
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFT6|A0A1L3KFT6_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 12 OX=1922805 PE=4 SV=1
MM1 pKa = 7.78PEE3 pKa = 3.67VSVTATGYY11 pKa = 10.14SKK13 pKa = 10.82KK14 pKa = 10.54APKK17 pKa = 9.73QVLSVLGLPSKK28 pKa = 10.47GQTWTHH34 pKa = 6.26ANDD37 pKa = 3.15VANVVEE43 pKa = 4.83SVHH46 pKa = 5.96EE47 pKa = 4.1RR48 pKa = 11.84VLGRR52 pKa = 11.84TADD55 pKa = 4.05GTWEE59 pKa = 3.71RR60 pKa = 11.84TMQPEE65 pKa = 4.04KK66 pKa = 11.0GAFTGDD72 pKa = 3.21LLVFRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84VGIHH83 pKa = 6.12TGSSSLPWSTEE94 pKa = 3.37QFVDD98 pKa = 4.14HH99 pKa = 6.69YY100 pKa = 10.65RR101 pKa = 11.84GQKK104 pKa = 7.92KK105 pKa = 9.78ARR107 pKa = 11.84YY108 pKa = 8.38QSAAKK113 pKa = 10.19SLEE116 pKa = 3.69EE117 pKa = 4.13RR118 pKa = 11.84PLSRR122 pKa = 11.84KK123 pKa = 9.75DD124 pKa = 3.4SYY126 pKa = 11.6PSVFLKK132 pKa = 10.69AEE134 pKa = 3.84KK135 pKa = 9.47WHH137 pKa = 6.71DD138 pKa = 3.71PKK140 pKa = 11.09AGRR143 pKa = 11.84LISARR148 pKa = 11.84HH149 pKa = 4.91PRR151 pKa = 11.84YY152 pKa = 9.92NLEE155 pKa = 4.03LGKK158 pKa = 10.61FILPLEE164 pKa = 4.48HH165 pKa = 6.9EE166 pKa = 4.74LYY168 pKa = 10.6KK169 pKa = 11.02AIDD172 pKa = 3.81EE173 pKa = 4.8VYY175 pKa = 10.92GSATIMKK182 pKa = 9.88GYY184 pKa = 8.78TPEE187 pKa = 3.41QRR189 pKa = 11.84AAVVEE194 pKa = 4.07QHH196 pKa = 5.82WGAFDD201 pKa = 3.71SPVAIGQDD209 pKa = 3.48YY210 pKa = 11.4SKK212 pKa = 10.95FDD214 pKa = 3.24QHH216 pKa = 7.35ISKK219 pKa = 10.63DD220 pKa = 3.35ALQYY224 pKa = 9.66EE225 pKa = 4.59HH226 pKa = 7.05GFYY229 pKa = 10.63LHH231 pKa = 7.47AYY233 pKa = 9.36GGDD236 pKa = 3.56EE237 pKa = 3.79NLQRR241 pKa = 11.84LLSWQLEE248 pKa = 4.4SRR250 pKa = 11.84CFANVQDD257 pKa = 4.87GVVQYY262 pKa = 8.67PVKK265 pKa = 9.93GGRR268 pKa = 11.84MSGDD272 pKa = 3.18MNTALGNCVISASLIWAYY290 pKa = 10.62ARR292 pKa = 11.84EE293 pKa = 4.16IGVKK297 pKa = 9.59IRR299 pKa = 11.84LVVDD303 pKa = 3.93GDD305 pKa = 3.95DD306 pKa = 3.37SVAFMEE312 pKa = 4.55RR313 pKa = 11.84TDD315 pKa = 3.4APRR318 pKa = 11.84YY319 pKa = 7.53QAGIKK324 pKa = 9.45EE325 pKa = 3.97WMARR329 pKa = 11.84RR330 pKa = 11.84GFRR333 pKa = 11.84LVSEE337 pKa = 4.37EE338 pKa = 3.84PVYY341 pKa = 10.7QINQVEE347 pKa = 4.99FCQCRR352 pKa = 11.84YY353 pKa = 10.58VGTTPPTMVRR363 pKa = 11.84NPLKK367 pKa = 10.77AITQDD372 pKa = 3.64HH373 pKa = 6.43AWVEE377 pKa = 4.55DD378 pKa = 3.57KK379 pKa = 11.0ALRR382 pKa = 11.84WADD385 pKa = 3.36VLAATGMGGLALYY398 pKa = 10.8GNIPLLGAYY407 pKa = 10.45YY408 pKa = 11.12DD409 pKa = 3.65MLARR413 pKa = 11.84ASEE416 pKa = 4.46VSAKK420 pKa = 9.49TLARR424 pKa = 11.84LDD426 pKa = 4.0TRR428 pKa = 11.84SSWLRR433 pKa = 11.84DD434 pKa = 3.2ATFGGVRR441 pKa = 11.84MEE443 pKa = 4.19PSEE446 pKa = 3.87MARR449 pKa = 11.84YY450 pKa = 8.8QFWLSWGMEE459 pKa = 3.71PGAQRR464 pKa = 11.84EE465 pKa = 4.25HH466 pKa = 6.48EE467 pKa = 4.16ARR469 pKa = 11.84FEE471 pKa = 4.63GIDD474 pKa = 3.78LSALKK479 pKa = 10.63AVDD482 pKa = 3.29TTKK485 pKa = 10.32IKK487 pKa = 10.64HH488 pKa = 5.59SSFYY492 pKa = 10.57DD493 pKa = 3.21TDD495 pKa = 3.07QAYY498 pKa = 10.74YY499 pKa = 9.97II500 pKa = 4.55
MM1 pKa = 7.78PEE3 pKa = 3.67VSVTATGYY11 pKa = 10.14SKK13 pKa = 10.82KK14 pKa = 10.54APKK17 pKa = 9.73QVLSVLGLPSKK28 pKa = 10.47GQTWTHH34 pKa = 6.26ANDD37 pKa = 3.15VANVVEE43 pKa = 4.83SVHH46 pKa = 5.96EE47 pKa = 4.1RR48 pKa = 11.84VLGRR52 pKa = 11.84TADD55 pKa = 4.05GTWEE59 pKa = 3.71RR60 pKa = 11.84TMQPEE65 pKa = 4.04KK66 pKa = 11.0GAFTGDD72 pKa = 3.21LLVFRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84VGIHH83 pKa = 6.12TGSSSLPWSTEE94 pKa = 3.37QFVDD98 pKa = 4.14HH99 pKa = 6.69YY100 pKa = 10.65RR101 pKa = 11.84GQKK104 pKa = 7.92KK105 pKa = 9.78ARR107 pKa = 11.84YY108 pKa = 8.38QSAAKK113 pKa = 10.19SLEE116 pKa = 3.69EE117 pKa = 4.13RR118 pKa = 11.84PLSRR122 pKa = 11.84KK123 pKa = 9.75DD124 pKa = 3.4SYY126 pKa = 11.6PSVFLKK132 pKa = 10.69AEE134 pKa = 3.84KK135 pKa = 9.47WHH137 pKa = 6.71DD138 pKa = 3.71PKK140 pKa = 11.09AGRR143 pKa = 11.84LISARR148 pKa = 11.84HH149 pKa = 4.91PRR151 pKa = 11.84YY152 pKa = 9.92NLEE155 pKa = 4.03LGKK158 pKa = 10.61FILPLEE164 pKa = 4.48HH165 pKa = 6.9EE166 pKa = 4.74LYY168 pKa = 10.6KK169 pKa = 11.02AIDD172 pKa = 3.81EE173 pKa = 4.8VYY175 pKa = 10.92GSATIMKK182 pKa = 9.88GYY184 pKa = 8.78TPEE187 pKa = 3.41QRR189 pKa = 11.84AAVVEE194 pKa = 4.07QHH196 pKa = 5.82WGAFDD201 pKa = 3.71SPVAIGQDD209 pKa = 3.48YY210 pKa = 11.4SKK212 pKa = 10.95FDD214 pKa = 3.24QHH216 pKa = 7.35ISKK219 pKa = 10.63DD220 pKa = 3.35ALQYY224 pKa = 9.66EE225 pKa = 4.59HH226 pKa = 7.05GFYY229 pKa = 10.63LHH231 pKa = 7.47AYY233 pKa = 9.36GGDD236 pKa = 3.56EE237 pKa = 3.79NLQRR241 pKa = 11.84LLSWQLEE248 pKa = 4.4SRR250 pKa = 11.84CFANVQDD257 pKa = 4.87GVVQYY262 pKa = 8.67PVKK265 pKa = 9.93GGRR268 pKa = 11.84MSGDD272 pKa = 3.18MNTALGNCVISASLIWAYY290 pKa = 10.62ARR292 pKa = 11.84EE293 pKa = 4.16IGVKK297 pKa = 9.59IRR299 pKa = 11.84LVVDD303 pKa = 3.93GDD305 pKa = 3.95DD306 pKa = 3.37SVAFMEE312 pKa = 4.55RR313 pKa = 11.84TDD315 pKa = 3.4APRR318 pKa = 11.84YY319 pKa = 7.53QAGIKK324 pKa = 9.45EE325 pKa = 3.97WMARR329 pKa = 11.84RR330 pKa = 11.84GFRR333 pKa = 11.84LVSEE337 pKa = 4.37EE338 pKa = 3.84PVYY341 pKa = 10.7QINQVEE347 pKa = 4.99FCQCRR352 pKa = 11.84YY353 pKa = 10.58VGTTPPTMVRR363 pKa = 11.84NPLKK367 pKa = 10.77AITQDD372 pKa = 3.64HH373 pKa = 6.43AWVEE377 pKa = 4.55DD378 pKa = 3.57KK379 pKa = 11.0ALRR382 pKa = 11.84WADD385 pKa = 3.36VLAATGMGGLALYY398 pKa = 10.8GNIPLLGAYY407 pKa = 10.45YY408 pKa = 11.12DD409 pKa = 3.65MLARR413 pKa = 11.84ASEE416 pKa = 4.46VSAKK420 pKa = 9.49TLARR424 pKa = 11.84LDD426 pKa = 4.0TRR428 pKa = 11.84SSWLRR433 pKa = 11.84DD434 pKa = 3.2ATFGGVRR441 pKa = 11.84MEE443 pKa = 4.19PSEE446 pKa = 3.87MARR449 pKa = 11.84YY450 pKa = 8.8QFWLSWGMEE459 pKa = 3.71PGAQRR464 pKa = 11.84EE465 pKa = 4.25HH466 pKa = 6.48EE467 pKa = 4.16ARR469 pKa = 11.84FEE471 pKa = 4.63GIDD474 pKa = 3.78LSALKK479 pKa = 10.63AVDD482 pKa = 3.29TTKK485 pKa = 10.32IKK487 pKa = 10.64HH488 pKa = 5.59SSFYY492 pKa = 10.57DD493 pKa = 3.21TDD495 pKa = 3.07QAYY498 pKa = 10.74YY499 pKa = 9.97II500 pKa = 4.55
Molecular weight: 56.37 kDa
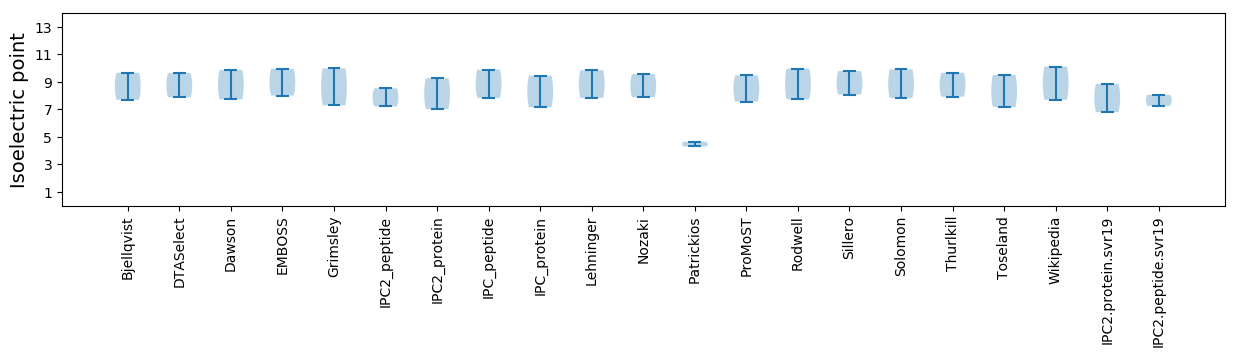
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFT6|A0A1L3KFT6_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 12 OX=1922805 PE=4 SV=1
MM1 pKa = 7.06VKK3 pKa = 10.15RR4 pKa = 11.84RR5 pKa = 11.84GSKK8 pKa = 10.17RR9 pKa = 11.84KK10 pKa = 7.64TNAPRR15 pKa = 11.84MRR17 pKa = 11.84NAPLAVRR24 pKa = 11.84APRR27 pKa = 11.84PRR29 pKa = 11.84MVSNEE34 pKa = 3.82EE35 pKa = 4.06KK36 pKa = 10.51LLKK39 pKa = 10.65LIMDD43 pKa = 4.46PCGAEE48 pKa = 3.96MAHH51 pKa = 6.88GYY53 pKa = 10.21ALSTEE58 pKa = 4.42GVVQRR63 pKa = 11.84FTQYY67 pKa = 7.69FTPTATTEE75 pKa = 3.86TSFAYY80 pKa = 10.15AINPAYY86 pKa = 10.06YY87 pKa = 10.55GPGGVNQVLGTGTGAIGTAISAPLPGQTFVDD118 pKa = 4.12ANADD122 pKa = 3.95TVSTLAACVEE132 pKa = 4.12VLYY135 pKa = 10.64TGTVVNRR142 pKa = 11.84KK143 pKa = 9.57GYY145 pKa = 9.7IAVAQGPSEE154 pKa = 4.2QIYY157 pKa = 8.2STITTPTVTFNGLVTLSNAVAPVPSAAVSVKK188 pKa = 7.43WTPPVANFIGSASAIEE204 pKa = 4.27TDD206 pKa = 2.86SHH208 pKa = 5.79YY209 pKa = 10.81QSNSILVTAVGVNPNDD225 pKa = 3.17FVVRR229 pKa = 11.84VTVVYY234 pKa = 9.85EE235 pKa = 3.72YY236 pKa = 11.13APKK239 pKa = 10.63VGLGMPLARR248 pKa = 11.84ATRR251 pKa = 11.84VTPVGVGEE259 pKa = 5.3RR260 pKa = 11.84ITSTLDD266 pKa = 3.34RR267 pKa = 11.84MGHH270 pKa = 4.67WWHH273 pKa = 6.5NLGDD277 pKa = 3.79AAAAASRR284 pKa = 11.84LGNRR288 pKa = 11.84VVYY291 pKa = 10.14GAGQVAGFTRR301 pKa = 11.84GLLRR305 pKa = 11.84TTEE308 pKa = 3.89AATTLLALAGG318 pKa = 3.87
MM1 pKa = 7.06VKK3 pKa = 10.15RR4 pKa = 11.84RR5 pKa = 11.84GSKK8 pKa = 10.17RR9 pKa = 11.84KK10 pKa = 7.64TNAPRR15 pKa = 11.84MRR17 pKa = 11.84NAPLAVRR24 pKa = 11.84APRR27 pKa = 11.84PRR29 pKa = 11.84MVSNEE34 pKa = 3.82EE35 pKa = 4.06KK36 pKa = 10.51LLKK39 pKa = 10.65LIMDD43 pKa = 4.46PCGAEE48 pKa = 3.96MAHH51 pKa = 6.88GYY53 pKa = 10.21ALSTEE58 pKa = 4.42GVVQRR63 pKa = 11.84FTQYY67 pKa = 7.69FTPTATTEE75 pKa = 3.86TSFAYY80 pKa = 10.15AINPAYY86 pKa = 10.06YY87 pKa = 10.55GPGGVNQVLGTGTGAIGTAISAPLPGQTFVDD118 pKa = 4.12ANADD122 pKa = 3.95TVSTLAACVEE132 pKa = 4.12VLYY135 pKa = 10.64TGTVVNRR142 pKa = 11.84KK143 pKa = 9.57GYY145 pKa = 9.7IAVAQGPSEE154 pKa = 4.2QIYY157 pKa = 8.2STITTPTVTFNGLVTLSNAVAPVPSAAVSVKK188 pKa = 7.43WTPPVANFIGSASAIEE204 pKa = 4.27TDD206 pKa = 2.86SHH208 pKa = 5.79YY209 pKa = 10.81QSNSILVTAVGVNPNDD225 pKa = 3.17FVVRR229 pKa = 11.84VTVVYY234 pKa = 9.85EE235 pKa = 3.72YY236 pKa = 11.13APKK239 pKa = 10.63VGLGMPLARR248 pKa = 11.84ATRR251 pKa = 11.84VTPVGVGEE259 pKa = 5.3RR260 pKa = 11.84ITSTLDD266 pKa = 3.34RR267 pKa = 11.84MGHH270 pKa = 4.67WWHH273 pKa = 6.5NLGDD277 pKa = 3.79AAAAASRR284 pKa = 11.84LGNRR288 pKa = 11.84VVYY291 pKa = 10.14GAGQVAGFTRR301 pKa = 11.84GLLRR305 pKa = 11.84TTEE308 pKa = 3.89AATTLLALAGG318 pKa = 3.87
Molecular weight: 33.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
818 |
318 |
500 |
409.0 |
44.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.002 ± 1.321 | 0.733 ± 0.063 |
4.279 ± 1.244 | 5.257 ± 1.077 |
2.934 ± 0.251 | 8.435 ± 0.598 |
2.2 ± 0.565 | 3.545 ± 0.052 |
4.156 ± 0.983 | 7.579 ± 0.396 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.445 ± 0.146 | 3.056 ± 0.995 |
5.012 ± 0.765 | 3.79 ± 0.763 |
6.601 ± 0.375 | 6.479 ± 0.49 |
7.335 ± 2.199 | 8.802 ± 1.509 |
1.956 ± 0.607 | 4.401 ± 0.376 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
