
BtVs-BetaCoV/SC2013
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Merbecovirus; Middle East respiratory syndrome-related coronavirus
Average proteome isoelectric point is 7.12
Get precalculated fractions of proteins
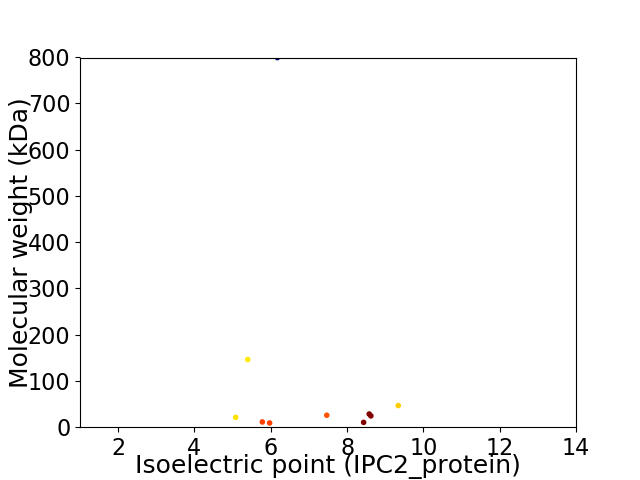
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A023YAI9|A0A023YAI9_MERS ORF4a OS=BtVs-BetaCoV/SC2013 OX=1495253 PE=4 SV=1
MM1 pKa = 6.72TTIPTLSSLEE11 pKa = 4.06AEE13 pKa = 4.5EE14 pKa = 4.71EE15 pKa = 4.16LLSLDD20 pKa = 4.06QLPITPSPGTLALPNTGRR38 pKa = 11.84SLFPFHH44 pKa = 7.69LDD46 pKa = 2.79RR47 pKa = 11.84AYY49 pKa = 11.36LLMPIQPLHH58 pKa = 6.44KK59 pKa = 9.24MLGIGGDD66 pKa = 3.19RR67 pKa = 11.84TEE69 pKa = 4.68RR70 pKa = 11.84LIQEE74 pKa = 4.42MEE76 pKa = 3.95QSSWLPGGSSTTLEE90 pKa = 4.6PGLKK94 pKa = 8.77LTSLSEE100 pKa = 4.06LSRR103 pKa = 11.84MEE105 pKa = 3.63SSGYY109 pKa = 7.78MKK111 pKa = 10.4KK112 pKa = 10.21VLQTLPPVLGRR123 pKa = 11.84VTLTTIRR130 pKa = 11.84LLLHH134 pKa = 6.83SSLQVLGFLKK144 pKa = 10.19TSTLRR149 pKa = 11.84ALEE152 pKa = 4.06ATVSHH157 pKa = 6.09LQEE160 pKa = 4.25HH161 pKa = 6.97LAPAEE166 pKa = 3.99ALQDD170 pKa = 3.79LAQEE174 pKa = 4.2VLVPEE179 pKa = 4.26THH181 pKa = 7.16PEE183 pKa = 3.91VLPQVQLEE191 pKa = 4.43SEE193 pKa = 4.34LL194 pKa = 3.84
MM1 pKa = 6.72TTIPTLSSLEE11 pKa = 4.06AEE13 pKa = 4.5EE14 pKa = 4.71EE15 pKa = 4.16LLSLDD20 pKa = 4.06QLPITPSPGTLALPNTGRR38 pKa = 11.84SLFPFHH44 pKa = 7.69LDD46 pKa = 2.79RR47 pKa = 11.84AYY49 pKa = 11.36LLMPIQPLHH58 pKa = 6.44KK59 pKa = 9.24MLGIGGDD66 pKa = 3.19RR67 pKa = 11.84TEE69 pKa = 4.68RR70 pKa = 11.84LIQEE74 pKa = 4.42MEE76 pKa = 3.95QSSWLPGGSSTTLEE90 pKa = 4.6PGLKK94 pKa = 8.77LTSLSEE100 pKa = 4.06LSRR103 pKa = 11.84MEE105 pKa = 3.63SSGYY109 pKa = 7.78MKK111 pKa = 10.4KK112 pKa = 10.21VLQTLPPVLGRR123 pKa = 11.84VTLTTIRR130 pKa = 11.84LLLHH134 pKa = 6.83SSLQVLGFLKK144 pKa = 10.19TSTLRR149 pKa = 11.84ALEE152 pKa = 4.06ATVSHH157 pKa = 6.09LQEE160 pKa = 4.25HH161 pKa = 6.97LAPAEE166 pKa = 3.99ALQDD170 pKa = 3.79LAQEE174 pKa = 4.2VLVPEE179 pKa = 4.26THH181 pKa = 7.16PEE183 pKa = 3.91VLPQVQLEE191 pKa = 4.43SEE193 pKa = 4.34LL194 pKa = 3.84
Molecular weight: 21.29 kDa
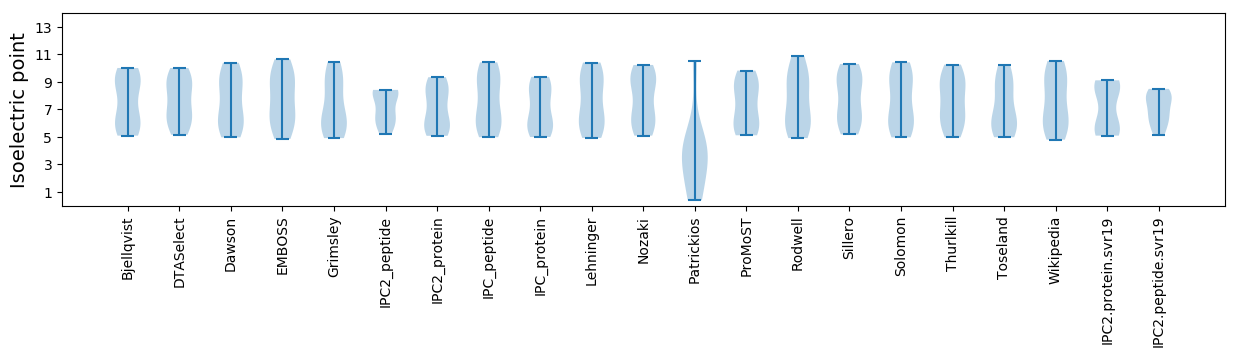
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A023YAJ4|A0A023YAJ4_MERS Nucleoprotein OS=BtVs-BetaCoV/SC2013 OX=1495253 GN=N PE=3 SV=1
MM1 pKa = 7.35ATPAAPRR8 pKa = 11.84AVTFGDD14 pKa = 3.57NNDD17 pKa = 3.83NNTNTQQSRR26 pKa = 11.84GRR28 pKa = 11.84GRR30 pKa = 11.84TPKK33 pKa = 9.93PRR35 pKa = 11.84PAPNNTVSWYY45 pKa = 9.65TGLTQHH51 pKa = 6.36GKK53 pKa = 9.97VPLSFPPGQGVPLNANSTPAQNAGYY78 pKa = 9.33WRR80 pKa = 11.84RR81 pKa = 11.84QDD83 pKa = 3.96RR84 pKa = 11.84KK85 pKa = 10.03INTGNGTKK93 pKa = 9.97QLAPRR98 pKa = 11.84WFFYY102 pKa = 9.92YY103 pKa = 10.27TGTGPEE109 pKa = 3.82ANLPFRR115 pKa = 11.84AVKK118 pKa = 10.55DD119 pKa = 4.02GIVWVHH125 pKa = 5.98EE126 pKa = 4.53EE127 pKa = 4.31GATDD131 pKa = 3.65APSSFGTRR139 pKa = 11.84NPNNDD144 pKa = 3.67PAIVTQFAPGTRR156 pKa = 11.84LPKK159 pKa = 10.18NFHH162 pKa = 6.51IEE164 pKa = 4.18GTGGNSQSSSRR175 pKa = 11.84ASSASRR181 pKa = 11.84GSSRR185 pKa = 11.84SSSRR189 pKa = 11.84GARR192 pKa = 11.84SGNSSRR198 pKa = 11.84SASPGPAGIGAVGGDD213 pKa = 3.62ASSILYY219 pKa = 10.5LSLLKK224 pKa = 10.62RR225 pKa = 11.84LEE227 pKa = 4.15DD228 pKa = 3.77LEE230 pKa = 4.75AGKK233 pKa = 10.6SKK235 pKa = 10.58SAPKK239 pKa = 10.58VVTKK243 pKa = 10.61KK244 pKa = 10.57DD245 pKa = 2.83AAAAKK250 pKa = 10.32NKK252 pKa = 8.56MRR254 pKa = 11.84HH255 pKa = 5.38KK256 pKa = 10.5RR257 pKa = 11.84VATKK261 pKa = 10.46SFNMVQAFGRR271 pKa = 11.84RR272 pKa = 11.84GPGDD276 pKa = 3.38LQGNFGDD283 pKa = 4.05LQLNKK288 pKa = 10.39LGTEE292 pKa = 4.19DD293 pKa = 4.25PRR295 pKa = 11.84WPQIAEE301 pKa = 4.2LAPSASAFMGMSQFKK316 pKa = 8.93LTHH319 pKa = 5.96QSNDD323 pKa = 3.01ADD325 pKa = 3.72GSPVYY330 pKa = 10.18FLRR333 pKa = 11.84YY334 pKa = 9.46SGAIKK339 pKa = 10.61LDD341 pKa = 3.7PKK343 pKa = 10.6NPNYY347 pKa = 9.39QKK349 pKa = 10.66WMEE352 pKa = 4.12LLEE355 pKa = 4.45ANIDD359 pKa = 3.68AYY361 pKa = 10.22KK362 pKa = 10.57SFPVKK367 pKa = 10.0EE368 pKa = 4.46KK369 pKa = 10.56KK370 pKa = 9.98QKK372 pKa = 11.13ADD374 pKa = 3.87DD375 pKa = 3.98NKK377 pKa = 10.54QDD379 pKa = 3.58STEE382 pKa = 4.13DD383 pKa = 3.52SGDD386 pKa = 3.53LFAEE390 pKa = 4.84AVASAMASQSLPQRR404 pKa = 11.84APKK407 pKa = 10.63GSITQRR413 pKa = 11.84SRR415 pKa = 11.84APRR418 pKa = 11.84APVSQMEE425 pKa = 4.86DD426 pKa = 3.1VNQTDD431 pKa = 3.8DD432 pKa = 3.56RR433 pKa = 11.84QQ434 pKa = 3.32
MM1 pKa = 7.35ATPAAPRR8 pKa = 11.84AVTFGDD14 pKa = 3.57NNDD17 pKa = 3.83NNTNTQQSRR26 pKa = 11.84GRR28 pKa = 11.84GRR30 pKa = 11.84TPKK33 pKa = 9.93PRR35 pKa = 11.84PAPNNTVSWYY45 pKa = 9.65TGLTQHH51 pKa = 6.36GKK53 pKa = 9.97VPLSFPPGQGVPLNANSTPAQNAGYY78 pKa = 9.33WRR80 pKa = 11.84RR81 pKa = 11.84QDD83 pKa = 3.96RR84 pKa = 11.84KK85 pKa = 10.03INTGNGTKK93 pKa = 9.97QLAPRR98 pKa = 11.84WFFYY102 pKa = 9.92YY103 pKa = 10.27TGTGPEE109 pKa = 3.82ANLPFRR115 pKa = 11.84AVKK118 pKa = 10.55DD119 pKa = 4.02GIVWVHH125 pKa = 5.98EE126 pKa = 4.53EE127 pKa = 4.31GATDD131 pKa = 3.65APSSFGTRR139 pKa = 11.84NPNNDD144 pKa = 3.67PAIVTQFAPGTRR156 pKa = 11.84LPKK159 pKa = 10.18NFHH162 pKa = 6.51IEE164 pKa = 4.18GTGGNSQSSSRR175 pKa = 11.84ASSASRR181 pKa = 11.84GSSRR185 pKa = 11.84SSSRR189 pKa = 11.84GARR192 pKa = 11.84SGNSSRR198 pKa = 11.84SASPGPAGIGAVGGDD213 pKa = 3.62ASSILYY219 pKa = 10.5LSLLKK224 pKa = 10.62RR225 pKa = 11.84LEE227 pKa = 4.15DD228 pKa = 3.77LEE230 pKa = 4.75AGKK233 pKa = 10.6SKK235 pKa = 10.58SAPKK239 pKa = 10.58VVTKK243 pKa = 10.61KK244 pKa = 10.57DD245 pKa = 2.83AAAAKK250 pKa = 10.32NKK252 pKa = 8.56MRR254 pKa = 11.84HH255 pKa = 5.38KK256 pKa = 10.5RR257 pKa = 11.84VATKK261 pKa = 10.46SFNMVQAFGRR271 pKa = 11.84RR272 pKa = 11.84GPGDD276 pKa = 3.38LQGNFGDD283 pKa = 4.05LQLNKK288 pKa = 10.39LGTEE292 pKa = 4.19DD293 pKa = 4.25PRR295 pKa = 11.84WPQIAEE301 pKa = 4.2LAPSASAFMGMSQFKK316 pKa = 8.93LTHH319 pKa = 5.96QSNDD323 pKa = 3.01ADD325 pKa = 3.72GSPVYY330 pKa = 10.18FLRR333 pKa = 11.84YY334 pKa = 9.46SGAIKK339 pKa = 10.61LDD341 pKa = 3.7PKK343 pKa = 10.6NPNYY347 pKa = 9.39QKK349 pKa = 10.66WMEE352 pKa = 4.12LLEE355 pKa = 4.45ANIDD359 pKa = 3.68AYY361 pKa = 10.22KK362 pKa = 10.57SFPVKK367 pKa = 10.0EE368 pKa = 4.46KK369 pKa = 10.56KK370 pKa = 9.98QKK372 pKa = 11.13ADD374 pKa = 3.87DD375 pKa = 3.98NKK377 pKa = 10.54QDD379 pKa = 3.58STEE382 pKa = 4.13DD383 pKa = 3.52SGDD386 pKa = 3.53LFAEE390 pKa = 4.84AVASAMASQSLPQRR404 pKa = 11.84APKK407 pKa = 10.63GSITQRR413 pKa = 11.84SRR415 pKa = 11.84APRR418 pKa = 11.84APVSQMEE425 pKa = 4.86DD426 pKa = 3.1VNQTDD431 pKa = 3.8DD432 pKa = 3.56RR433 pKa = 11.84QQ434 pKa = 3.32
Molecular weight: 46.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10106 |
82 |
7179 |
1010.6 |
112.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.896 ± 0.451 | 3.107 ± 0.554 |
5.333 ± 0.51 | 3.948 ± 0.298 |
4.938 ± 0.372 | 5.897 ± 0.42 |
2.058 ± 0.375 | 4.453 ± 0.441 |
5.274 ± 0.684 | 9.321 ± 0.659 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.187 ± 0.132 | 5.096 ± 0.343 |
4.532 ± 0.801 | 3.82 ± 0.712 |
3.859 ± 0.543 | 7.263 ± 0.709 |
6.471 ± 0.193 | 8.698 ± 0.772 |
1.118 ± 0.217 | 4.73 ± 0.512 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
