
Sugarcane white streak virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
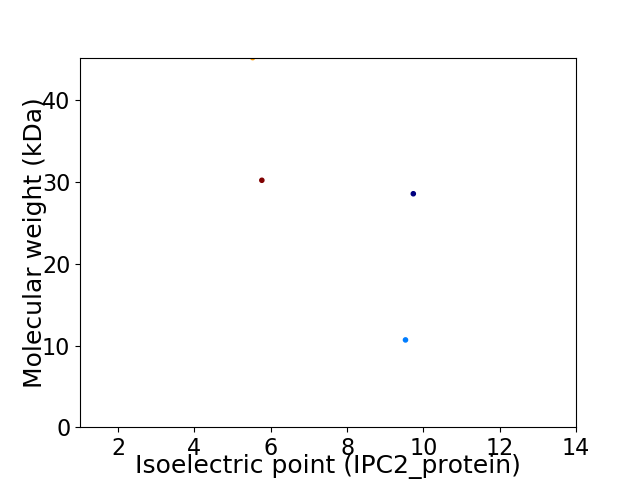
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|X5CJJ1|X5CJJ1_9GEMI Replication-associated protein OS=Sugarcane white streak virus OX=1492296 PE=3 SV=1
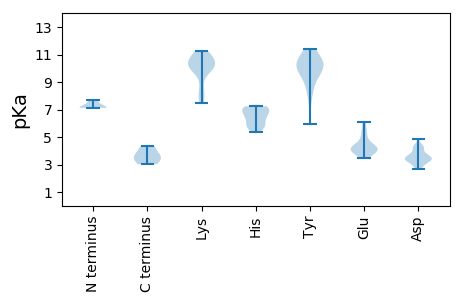
MM1 pKa = 7.12STDD4 pKa = 3.37SSNQGIAAQRR14 pKa = 11.84LSPGLEE20 pKa = 3.98EE21 pKa = 5.47FTTSWPDD28 pKa = 3.11PRR30 pKa = 11.84AGSTSQPRR38 pKa = 11.84VFQFKK43 pKa = 10.01AQNIFLTYY51 pKa = 8.57PRR53 pKa = 11.84CDD55 pKa = 2.95ISVDD59 pKa = 3.38VAARR63 pKa = 11.84TLLTLCHH70 pKa = 6.94RR71 pKa = 11.84FQPLYY76 pKa = 9.76ILCSQEE82 pKa = 3.54HH83 pKa = 6.34HH84 pKa = 7.11ADD86 pKa = 3.66GSNHH90 pKa = 5.74LHH92 pKa = 7.08ILLQTDD98 pKa = 2.66KK99 pKa = 11.23TMYY102 pKa = 9.06TRR104 pKa = 11.84NPHH107 pKa = 5.75YY108 pKa = 10.61FDD110 pKa = 3.64ICGHH114 pKa = 6.5HH115 pKa = 7.23PNIQPAKK122 pKa = 10.16SPDD125 pKa = 3.26NVRR128 pKa = 11.84AYY130 pKa = 9.73ILKK133 pKa = 10.55DD134 pKa = 3.64PITSFEE140 pKa = 4.25EE141 pKa = 5.17GSFQPRR147 pKa = 11.84GSRR150 pKa = 11.84SNTSAIPRR158 pKa = 11.84SGNSGTKK165 pKa = 9.95DD166 pKa = 2.96SLMRR170 pKa = 11.84DD171 pKa = 3.99IINTSTSKK179 pKa = 11.0DD180 pKa = 3.43DD181 pKa = 3.36YY182 pKa = 10.09LTRR185 pKa = 11.84VRR187 pKa = 11.84NTFPFDD193 pKa = 2.74WATRR197 pKa = 11.84LQQFEE202 pKa = 4.32YY203 pKa = 10.29SASKK207 pKa = 10.56LFPEE211 pKa = 4.71PVRR214 pKa = 11.84EE215 pKa = 4.09YY216 pKa = 11.38VNPFPPSEE224 pKa = 4.4PDD226 pKa = 3.33LFCRR230 pKa = 11.84EE231 pKa = 4.09IIDD234 pKa = 3.23RR235 pKa = 11.84WVYY238 pKa = 11.35DD239 pKa = 4.32EE240 pKa = 6.07LDD242 pKa = 3.38MDD244 pKa = 3.53ITDD247 pKa = 4.87AFDD250 pKa = 3.37AAQRR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84SLYY259 pKa = 9.46IVGPTRR265 pKa = 11.84TGKK268 pKa = 8.25STWARR273 pKa = 11.84SLGRR277 pKa = 11.84HH278 pKa = 5.9NYY280 pKa = 5.92WQHH283 pKa = 5.35MVDD286 pKa = 4.24FTAYY290 pKa = 7.98DD291 pKa = 3.45TLAKK295 pKa = 10.76YY296 pKa = 10.66NILDD300 pKa = 3.74DD301 pKa = 4.38VPFKK305 pKa = 10.89FCPNWKK311 pKa = 9.47QLVGCQRR318 pKa = 11.84DD319 pKa = 4.19FIVNPKK325 pKa = 7.51YY326 pKa = 10.72AKK328 pKa = 10.08RR329 pKa = 11.84KK330 pKa = 7.85EE331 pKa = 4.07IPGGIPCIILQNPDD345 pKa = 4.37DD346 pKa = 4.28DD347 pKa = 4.39WLPVLSPSQMDD358 pKa = 3.6YY359 pKa = 11.01FVNNCDD365 pKa = 3.53VYY367 pKa = 11.31VMKK370 pKa = 10.46PGEE373 pKa = 4.24KK374 pKa = 10.03FFGGDD379 pKa = 3.28TPVPEE384 pKa = 4.48AQEE387 pKa = 4.21DD388 pKa = 4.45VPDD391 pKa = 4.1GTGSSS396 pKa = 3.61
MM1 pKa = 7.12STDD4 pKa = 3.37SSNQGIAAQRR14 pKa = 11.84LSPGLEE20 pKa = 3.98EE21 pKa = 5.47FTTSWPDD28 pKa = 3.11PRR30 pKa = 11.84AGSTSQPRR38 pKa = 11.84VFQFKK43 pKa = 10.01AQNIFLTYY51 pKa = 8.57PRR53 pKa = 11.84CDD55 pKa = 2.95ISVDD59 pKa = 3.38VAARR63 pKa = 11.84TLLTLCHH70 pKa = 6.94RR71 pKa = 11.84FQPLYY76 pKa = 9.76ILCSQEE82 pKa = 3.54HH83 pKa = 6.34HH84 pKa = 7.11ADD86 pKa = 3.66GSNHH90 pKa = 5.74LHH92 pKa = 7.08ILLQTDD98 pKa = 2.66KK99 pKa = 11.23TMYY102 pKa = 9.06TRR104 pKa = 11.84NPHH107 pKa = 5.75YY108 pKa = 10.61FDD110 pKa = 3.64ICGHH114 pKa = 6.5HH115 pKa = 7.23PNIQPAKK122 pKa = 10.16SPDD125 pKa = 3.26NVRR128 pKa = 11.84AYY130 pKa = 9.73ILKK133 pKa = 10.55DD134 pKa = 3.64PITSFEE140 pKa = 4.25EE141 pKa = 5.17GSFQPRR147 pKa = 11.84GSRR150 pKa = 11.84SNTSAIPRR158 pKa = 11.84SGNSGTKK165 pKa = 9.95DD166 pKa = 2.96SLMRR170 pKa = 11.84DD171 pKa = 3.99IINTSTSKK179 pKa = 11.0DD180 pKa = 3.43DD181 pKa = 3.36YY182 pKa = 10.09LTRR185 pKa = 11.84VRR187 pKa = 11.84NTFPFDD193 pKa = 2.74WATRR197 pKa = 11.84LQQFEE202 pKa = 4.32YY203 pKa = 10.29SASKK207 pKa = 10.56LFPEE211 pKa = 4.71PVRR214 pKa = 11.84EE215 pKa = 4.09YY216 pKa = 11.38VNPFPPSEE224 pKa = 4.4PDD226 pKa = 3.33LFCRR230 pKa = 11.84EE231 pKa = 4.09IIDD234 pKa = 3.23RR235 pKa = 11.84WVYY238 pKa = 11.35DD239 pKa = 4.32EE240 pKa = 6.07LDD242 pKa = 3.38MDD244 pKa = 3.53ITDD247 pKa = 4.87AFDD250 pKa = 3.37AAQRR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84SLYY259 pKa = 9.46IVGPTRR265 pKa = 11.84TGKK268 pKa = 8.25STWARR273 pKa = 11.84SLGRR277 pKa = 11.84HH278 pKa = 5.9NYY280 pKa = 5.92WQHH283 pKa = 5.35MVDD286 pKa = 4.24FTAYY290 pKa = 7.98DD291 pKa = 3.45TLAKK295 pKa = 10.76YY296 pKa = 10.66NILDD300 pKa = 3.74DD301 pKa = 4.38VPFKK305 pKa = 10.89FCPNWKK311 pKa = 9.47QLVGCQRR318 pKa = 11.84DD319 pKa = 4.19FIVNPKK325 pKa = 7.51YY326 pKa = 10.72AKK328 pKa = 10.08RR329 pKa = 11.84KK330 pKa = 7.85EE331 pKa = 4.07IPGGIPCIILQNPDD345 pKa = 4.37DD346 pKa = 4.28DD347 pKa = 4.39WLPVLSPSQMDD358 pKa = 3.6YY359 pKa = 11.01FVNNCDD365 pKa = 3.53VYY367 pKa = 11.31VMKK370 pKa = 10.46PGEE373 pKa = 4.24KK374 pKa = 10.03FFGGDD379 pKa = 3.28TPVPEE384 pKa = 4.48AQEE387 pKa = 4.21DD388 pKa = 4.45VPDD391 pKa = 4.1GTGSSS396 pKa = 3.61
Molecular weight: 45.25 kDa
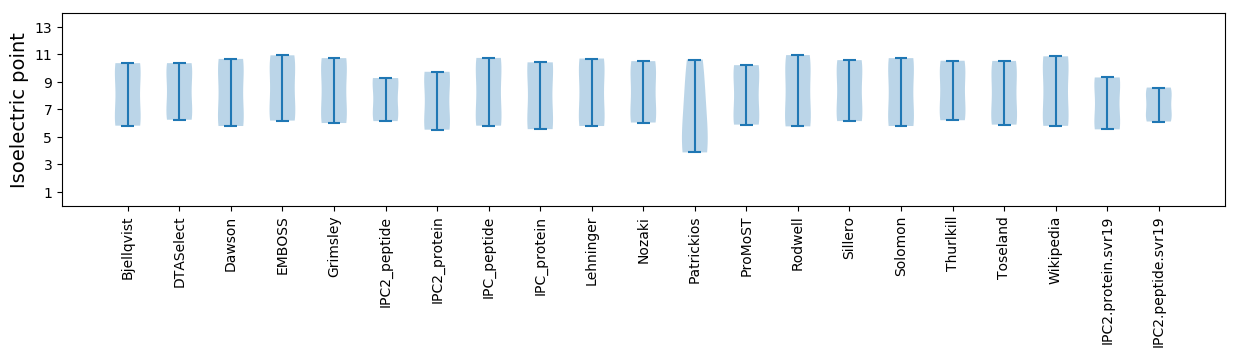
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X5CZY0|X5CZY0_9GEMI Capsid protein OS=Sugarcane white streak virus OX=1492296 PE=3 SV=1
MM1 pKa = 7.72AGPMRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 8.98PARR11 pKa = 11.84SSRR14 pKa = 11.84RR15 pKa = 11.84IVPWPAEE22 pKa = 3.79ALRR25 pKa = 11.84RR26 pKa = 11.84GYY28 pKa = 8.29TPRR31 pKa = 11.84NSVLRR36 pKa = 11.84GTRR39 pKa = 11.84SGRR42 pKa = 11.84SDD44 pKa = 3.46RR45 pKa = 11.84RR46 pKa = 11.84PSLQVINYY54 pKa = 7.44RR55 pKa = 11.84WSGQSAPIFKK65 pKa = 9.86TGGATYY71 pKa = 10.55LSSTFARR78 pKa = 11.84GNGEE82 pKa = 3.81DD83 pKa = 3.41QRR85 pKa = 11.84RR86 pKa = 11.84GAEE89 pKa = 3.96TIIYY93 pKa = 9.59KK94 pKa = 10.09IAFKK98 pKa = 10.85LVFAVTSSQLKK109 pKa = 8.11YY110 pKa = 10.57VSRR113 pKa = 11.84GMCVLWLVYY122 pKa = 9.95DD123 pKa = 4.3AQPTGIKK130 pKa = 9.87PEE132 pKa = 4.21PKK134 pKa = 10.22DD135 pKa = 3.1IFDD138 pKa = 4.34YY139 pKa = 9.6DD140 pKa = 3.72TGLSQSPMTWSVQRR154 pKa = 11.84ALCNRR159 pKa = 11.84FVVKK163 pKa = 10.42RR164 pKa = 11.84RR165 pKa = 11.84WKK167 pKa = 10.81FEE169 pKa = 3.82LSVNGVSAADD179 pKa = 4.14DD180 pKa = 3.64SSDD183 pKa = 3.34AQLSGKK189 pKa = 7.61TIPYY193 pKa = 9.25SRR195 pKa = 11.84RR196 pKa = 11.84GFDD199 pKa = 3.35KK200 pKa = 10.24FCKK203 pKa = 10.0RR204 pKa = 11.84LATRR208 pKa = 11.84TEE210 pKa = 3.82WKK212 pKa = 8.49NTEE215 pKa = 4.14GGDD218 pKa = 3.86IGDD221 pKa = 3.81IQKK224 pKa = 10.89GALYY228 pKa = 10.58VIAAPGNAMEE238 pKa = 4.8FQAIGNIRR246 pKa = 11.84VYY248 pKa = 10.24FKK250 pKa = 11.13SVGLQQ255 pKa = 3.03
MM1 pKa = 7.72AGPMRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 8.98PARR11 pKa = 11.84SSRR14 pKa = 11.84RR15 pKa = 11.84IVPWPAEE22 pKa = 3.79ALRR25 pKa = 11.84RR26 pKa = 11.84GYY28 pKa = 8.29TPRR31 pKa = 11.84NSVLRR36 pKa = 11.84GTRR39 pKa = 11.84SGRR42 pKa = 11.84SDD44 pKa = 3.46RR45 pKa = 11.84RR46 pKa = 11.84PSLQVINYY54 pKa = 7.44RR55 pKa = 11.84WSGQSAPIFKK65 pKa = 9.86TGGATYY71 pKa = 10.55LSSTFARR78 pKa = 11.84GNGEE82 pKa = 3.81DD83 pKa = 3.41QRR85 pKa = 11.84RR86 pKa = 11.84GAEE89 pKa = 3.96TIIYY93 pKa = 9.59KK94 pKa = 10.09IAFKK98 pKa = 10.85LVFAVTSSQLKK109 pKa = 8.11YY110 pKa = 10.57VSRR113 pKa = 11.84GMCVLWLVYY122 pKa = 9.95DD123 pKa = 4.3AQPTGIKK130 pKa = 9.87PEE132 pKa = 4.21PKK134 pKa = 10.22DD135 pKa = 3.1IFDD138 pKa = 4.34YY139 pKa = 9.6DD140 pKa = 3.72TGLSQSPMTWSVQRR154 pKa = 11.84ALCNRR159 pKa = 11.84FVVKK163 pKa = 10.42RR164 pKa = 11.84RR165 pKa = 11.84WKK167 pKa = 10.81FEE169 pKa = 3.82LSVNGVSAADD179 pKa = 4.14DD180 pKa = 3.64SSDD183 pKa = 3.34AQLSGKK189 pKa = 7.61TIPYY193 pKa = 9.25SRR195 pKa = 11.84RR196 pKa = 11.84GFDD199 pKa = 3.35KK200 pKa = 10.24FCKK203 pKa = 10.0RR204 pKa = 11.84LATRR208 pKa = 11.84TEE210 pKa = 3.82WKK212 pKa = 8.49NTEE215 pKa = 4.14GGDD218 pKa = 3.86IGDD221 pKa = 3.81IQKK224 pKa = 10.89GALYY228 pKa = 10.58VIAAPGNAMEE238 pKa = 4.8FQAIGNIRR246 pKa = 11.84VYY248 pKa = 10.24FKK250 pKa = 11.13SVGLQQ255 pKa = 3.03
Molecular weight: 28.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1012 |
97 |
396 |
253.0 |
28.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.83 ± 0.708 | 1.779 ± 0.239 |
6.719 ± 0.909 | 3.557 ± 0.255 |
5.138 ± 0.301 | 6.126 ± 1.049 |
2.174 ± 0.676 | 5.83 ± 0.236 |
3.854 ± 0.747 | 6.818 ± 0.421 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.779 ± 0.087 | 3.854 ± 0.363 |
7.411 ± 0.693 | 4.447 ± 0.204 |
7.905 ± 0.82 | 8.893 ± 0.791 |
6.423 ± 0.288 | 5.632 ± 1.064 |
1.779 ± 0.228 | 4.051 ± 0.164 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
