
Human papillomavirus 135
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 15
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
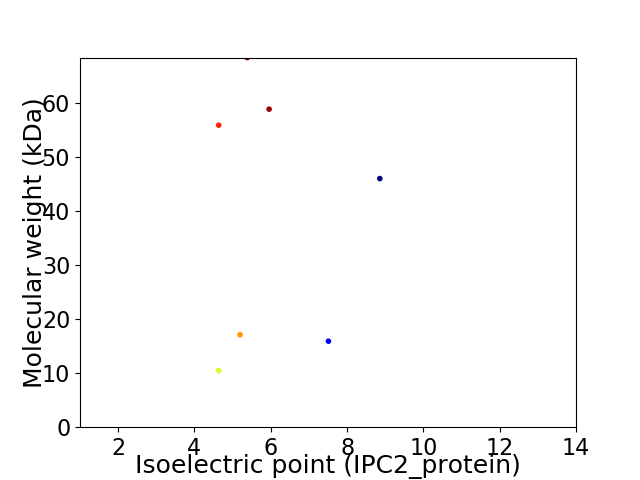
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
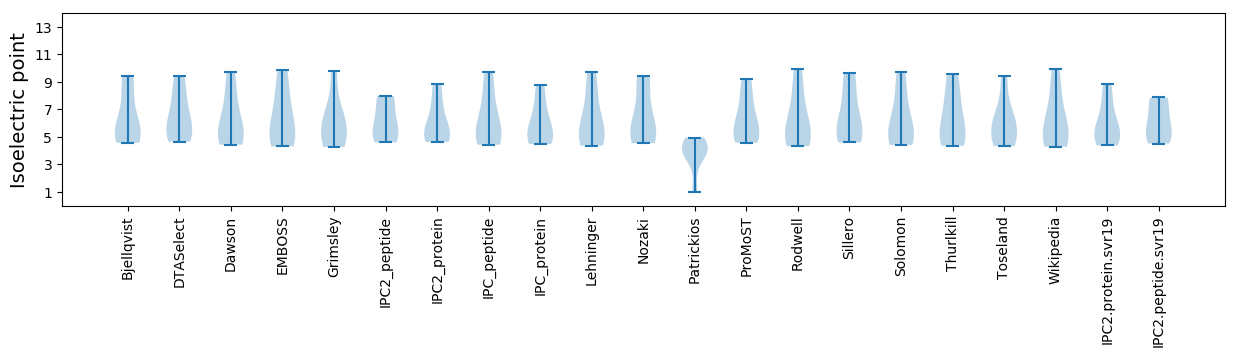
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3P6K5|I3P6K5_9PAPI Replication protein E1 OS=Human papillomavirus 135 OX=1070408 GN=E1 PE=3 SV=1
MM1 pKa = 7.69KK2 pKa = 9.47GTIATIPDD10 pKa = 3.29VCLEE14 pKa = 3.93EE15 pKa = 5.32LVLPSNLLASEE26 pKa = 4.62EE27 pKa = 4.29SLSPDD32 pKa = 3.85DD33 pKa = 4.51EE34 pKa = 5.28PEE36 pKa = 4.04EE37 pKa = 4.65EE38 pKa = 4.17PTNPYY43 pKa = 10.29RR44 pKa = 11.84VDD46 pKa = 4.0TYY48 pKa = 11.13CGNCQRR54 pKa = 11.84GVRR57 pKa = 11.84LFFVTTASCIRR68 pKa = 11.84TVHH71 pKa = 6.27HH72 pKa = 6.58LLIGEE77 pKa = 4.5LSVICVACSRR87 pKa = 11.84TCFQDD92 pKa = 3.18GRR94 pKa = 11.84PP95 pKa = 3.57
MM1 pKa = 7.69KK2 pKa = 9.47GTIATIPDD10 pKa = 3.29VCLEE14 pKa = 3.93EE15 pKa = 5.32LVLPSNLLASEE26 pKa = 4.62EE27 pKa = 4.29SLSPDD32 pKa = 3.85DD33 pKa = 4.51EE34 pKa = 5.28PEE36 pKa = 4.04EE37 pKa = 4.65EE38 pKa = 4.17PTNPYY43 pKa = 10.29RR44 pKa = 11.84VDD46 pKa = 4.0TYY48 pKa = 11.13CGNCQRR54 pKa = 11.84GVRR57 pKa = 11.84LFFVTTASCIRR68 pKa = 11.84TVHH71 pKa = 6.27HH72 pKa = 6.58LLIGEE77 pKa = 4.5LSVICVACSRR87 pKa = 11.84TCFQDD92 pKa = 3.18GRR94 pKa = 11.84PP95 pKa = 3.57
Molecular weight: 10.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3P6K7|I3P6K7_9PAPI Uncharacterized protein OS=Human papillomavirus 135 OX=1070408 GN=E4 PE=4 SV=1
MM1 pKa = 7.0NQADD5 pKa = 3.92LTDD8 pKa = 4.26RR9 pKa = 11.84FDD11 pKa = 3.65VLQDD15 pKa = 3.38QLLALYY21 pKa = 9.42EE22 pKa = 4.27KK23 pKa = 10.73GATDD27 pKa = 4.03LASQIKK33 pKa = 9.35HH34 pKa = 5.52WEE36 pKa = 3.82LSRR39 pKa = 11.84KK40 pKa = 9.64INVLMYY46 pKa = 8.01YY47 pKa = 9.85SRR49 pKa = 11.84KK50 pKa = 9.64EE51 pKa = 3.66GHH53 pKa = 6.77KK54 pKa = 10.72NLGLQTLPTLQVSEE68 pKa = 4.71YY69 pKa = 9.64NAKK72 pKa = 10.25LAIKK76 pKa = 9.88MMILLKK82 pKa = 10.75SLASSKK88 pKa = 10.51YY89 pKa = 9.91GKK91 pKa = 10.08EE92 pKa = 3.44PWTLTEE98 pKa = 4.12TSADD102 pKa = 3.85LLLTPPKK109 pKa = 9.96NTFKK113 pKa = 10.96KK114 pKa = 10.47GGFQVEE120 pKa = 4.45VYY122 pKa = 10.78YY123 pKa = 11.17DD124 pKa = 4.0NDD126 pKa = 3.67PQNANVYY133 pKa = 6.84TQWEE137 pKa = 4.04FLYY140 pKa = 10.98YY141 pKa = 9.99QDD143 pKa = 6.34LNDD146 pKa = 3.84EE147 pKa = 4.2WHH149 pKa = 6.67KK150 pKa = 11.27VPGDD154 pKa = 3.42VDD156 pKa = 4.42HH157 pKa = 7.16NGLSYY162 pKa = 11.26TDD164 pKa = 3.18ITGEE168 pKa = 3.95KK169 pKa = 9.06IYY171 pKa = 10.55FKK173 pKa = 10.47IFSEE177 pKa = 4.66DD178 pKa = 3.0ADD180 pKa = 4.11RR181 pKa = 11.84YY182 pKa = 10.51SNTGQWTVKK191 pKa = 10.2FKK193 pKa = 10.42STTISSVVTSSKK205 pKa = 10.81KK206 pKa = 10.45SFSAEE211 pKa = 3.83KK212 pKa = 10.5GDD214 pKa = 3.92SQRR217 pKa = 11.84SQHH220 pKa = 6.17RR221 pKa = 11.84EE222 pKa = 3.58SSPEE226 pKa = 3.76EE227 pKa = 4.25GTSSRR232 pKa = 11.84DD233 pKa = 3.14PRR235 pKa = 11.84RR236 pKa = 11.84SKK238 pKa = 10.53RR239 pKa = 11.84YY240 pKa = 8.73SQEE243 pKa = 3.84EE244 pKa = 4.58LPTTTTASPTTSTRR258 pKa = 11.84EE259 pKa = 3.33RR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84GSGGEE270 pKa = 3.86QQGEE274 pKa = 4.46STTRR278 pKa = 11.84EE279 pKa = 3.69PRR281 pKa = 11.84AKK283 pKa = 9.47RR284 pKa = 11.84TRR286 pKa = 11.84GPTAPTPDD294 pKa = 3.33QVGSRR299 pKa = 11.84HH300 pKa = 6.07RR301 pKa = 11.84SVPSHH306 pKa = 5.43NLGRR310 pKa = 11.84LGRR313 pKa = 11.84LQADD317 pKa = 3.07AWDD320 pKa = 4.12PPILCVKK327 pKa = 10.62GPANNLKK334 pKa = 10.12CWRR337 pKa = 11.84NRR339 pKa = 11.84FNVKK343 pKa = 9.47FHH345 pKa = 6.83SLFFNVSSVFKK356 pKa = 10.57WLGDD360 pKa = 3.65SNADD364 pKa = 3.57HH365 pKa = 6.97NISRR369 pKa = 11.84MLIAFHH375 pKa = 6.73DD376 pKa = 3.96TYY378 pKa = 11.37EE379 pKa = 4.16RR380 pKa = 11.84ARR382 pKa = 11.84FLQTVTLPRR391 pKa = 11.84GATYY395 pKa = 10.18AYY397 pKa = 10.44GSLDD401 pKa = 3.38SLL403 pKa = 4.41
MM1 pKa = 7.0NQADD5 pKa = 3.92LTDD8 pKa = 4.26RR9 pKa = 11.84FDD11 pKa = 3.65VLQDD15 pKa = 3.38QLLALYY21 pKa = 9.42EE22 pKa = 4.27KK23 pKa = 10.73GATDD27 pKa = 4.03LASQIKK33 pKa = 9.35HH34 pKa = 5.52WEE36 pKa = 3.82LSRR39 pKa = 11.84KK40 pKa = 9.64INVLMYY46 pKa = 8.01YY47 pKa = 9.85SRR49 pKa = 11.84KK50 pKa = 9.64EE51 pKa = 3.66GHH53 pKa = 6.77KK54 pKa = 10.72NLGLQTLPTLQVSEE68 pKa = 4.71YY69 pKa = 9.64NAKK72 pKa = 10.25LAIKK76 pKa = 9.88MMILLKK82 pKa = 10.75SLASSKK88 pKa = 10.51YY89 pKa = 9.91GKK91 pKa = 10.08EE92 pKa = 3.44PWTLTEE98 pKa = 4.12TSADD102 pKa = 3.85LLLTPPKK109 pKa = 9.96NTFKK113 pKa = 10.96KK114 pKa = 10.47GGFQVEE120 pKa = 4.45VYY122 pKa = 10.78YY123 pKa = 11.17DD124 pKa = 4.0NDD126 pKa = 3.67PQNANVYY133 pKa = 6.84TQWEE137 pKa = 4.04FLYY140 pKa = 10.98YY141 pKa = 9.99QDD143 pKa = 6.34LNDD146 pKa = 3.84EE147 pKa = 4.2WHH149 pKa = 6.67KK150 pKa = 11.27VPGDD154 pKa = 3.42VDD156 pKa = 4.42HH157 pKa = 7.16NGLSYY162 pKa = 11.26TDD164 pKa = 3.18ITGEE168 pKa = 3.95KK169 pKa = 9.06IYY171 pKa = 10.55FKK173 pKa = 10.47IFSEE177 pKa = 4.66DD178 pKa = 3.0ADD180 pKa = 4.11RR181 pKa = 11.84YY182 pKa = 10.51SNTGQWTVKK191 pKa = 10.2FKK193 pKa = 10.42STTISSVVTSSKK205 pKa = 10.81KK206 pKa = 10.45SFSAEE211 pKa = 3.83KK212 pKa = 10.5GDD214 pKa = 3.92SQRR217 pKa = 11.84SQHH220 pKa = 6.17RR221 pKa = 11.84EE222 pKa = 3.58SSPEE226 pKa = 3.76EE227 pKa = 4.25GTSSRR232 pKa = 11.84DD233 pKa = 3.14PRR235 pKa = 11.84RR236 pKa = 11.84SKK238 pKa = 10.53RR239 pKa = 11.84YY240 pKa = 8.73SQEE243 pKa = 3.84EE244 pKa = 4.58LPTTTTASPTTSTRR258 pKa = 11.84EE259 pKa = 3.33RR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84GSGGEE270 pKa = 3.86QQGEE274 pKa = 4.46STTRR278 pKa = 11.84EE279 pKa = 3.69PRR281 pKa = 11.84AKK283 pKa = 9.47RR284 pKa = 11.84TRR286 pKa = 11.84GPTAPTPDD294 pKa = 3.33QVGSRR299 pKa = 11.84HH300 pKa = 6.07RR301 pKa = 11.84SVPSHH306 pKa = 5.43NLGRR310 pKa = 11.84LGRR313 pKa = 11.84LQADD317 pKa = 3.07AWDD320 pKa = 4.12PPILCVKK327 pKa = 10.62GPANNLKK334 pKa = 10.12CWRR337 pKa = 11.84NRR339 pKa = 11.84FNVKK343 pKa = 9.47FHH345 pKa = 6.83SLFFNVSSVFKK356 pKa = 10.57WLGDD360 pKa = 3.65SNADD364 pKa = 3.57HH365 pKa = 6.97NISRR369 pKa = 11.84MLIAFHH375 pKa = 6.73DD376 pKa = 3.96TYY378 pKa = 11.37EE379 pKa = 4.16RR380 pKa = 11.84ARR382 pKa = 11.84FLQTVTLPRR391 pKa = 11.84GATYY395 pKa = 10.18AYY397 pKa = 10.44GSLDD401 pKa = 3.38SLL403 pKa = 4.41
Molecular weight: 46.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2412 |
95 |
601 |
344.6 |
38.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.39 ± 0.642 | 1.99 ± 0.778 |
7.09 ± 0.316 | 5.556 ± 0.366 |
4.726 ± 0.387 | 5.473 ± 0.811 |
2.114 ± 0.212 | 5.1 ± 0.676 |
5.556 ± 0.669 | 8.955 ± 0.777 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.493 ± 0.372 | 5.141 ± 0.641 |
6.343 ± 1.098 | 4.312 ± 0.398 |
5.597 ± 0.536 | 7.338 ± 0.664 |
6.758 ± 0.882 | 6.219 ± 0.68 |
1.161 ± 0.328 | 3.69 ± 0.368 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
