
Arcobacter sp. CECT 8987
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Campylobacteraceae; Arcobacter group; unclassified Arcobacter group
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
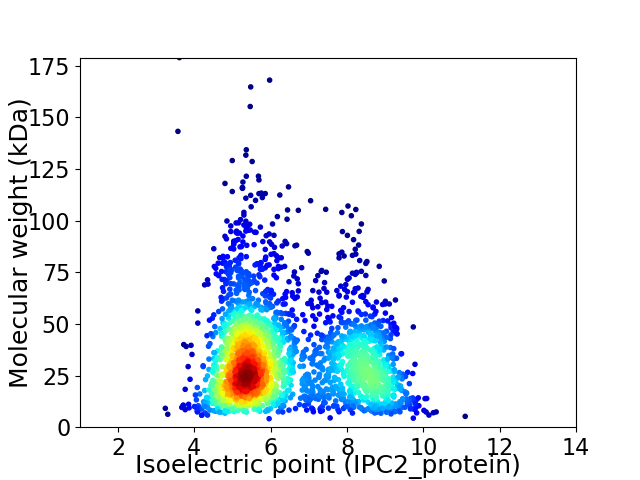
Virtual 2D-PAGE plot for 2486 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q0XQN1|A0A4Q0XQN1_9PROT Uncharacterized protein OS=Arcobacter sp. CECT 8987 OX=2044504 GN=CRV04_06160 PE=4 SV=1
MM1 pKa = 7.67AEE3 pKa = 4.54SIAKK7 pKa = 8.93VQSLLGKK14 pKa = 9.84FYY16 pKa = 11.11AQDD19 pKa = 2.98EE20 pKa = 4.73SGNVRR25 pKa = 11.84VLKK28 pKa = 11.02VGDD31 pKa = 3.75TIYY34 pKa = 10.57QGEE37 pKa = 4.57KK38 pKa = 10.47VYY40 pKa = 11.35GDD42 pKa = 3.7AANNSSDD49 pKa = 3.21KK50 pKa = 11.09LQIVSDD56 pKa = 3.83TQEE59 pKa = 4.0TFDD62 pKa = 3.63ITGSDD67 pKa = 3.36SLVFDD72 pKa = 4.23TSTNEE77 pKa = 3.58ATFGNEE83 pKa = 2.93EE84 pKa = 4.07VAFNPNSAWMPTADD98 pKa = 3.42NATVPEE104 pKa = 4.79GEE106 pKa = 4.68TVTDD110 pKa = 3.71AQGDD114 pKa = 3.65ITEE117 pKa = 4.65EE118 pKa = 3.63EE119 pKa = 4.11TAAGNEE125 pKa = 4.17AASSSDD131 pKa = 2.99VGVFNFEE138 pKa = 3.97NRR140 pKa = 11.84DD141 pKa = 3.35GDD143 pKa = 4.25SVNIQSDD150 pKa = 3.27LRR152 pKa = 11.84EE153 pKa = 4.19SVFSNNSDD161 pKa = 3.28SLVSNNGDD169 pKa = 3.43GLNSNPSSNLLLNGTPTPDD188 pKa = 3.65DD189 pKa = 4.12VNDD192 pKa = 3.52VPVAVDD198 pKa = 4.61DD199 pKa = 5.16SFTLDD204 pKa = 3.21EE205 pKa = 4.74DD206 pKa = 3.78ASYY209 pKa = 10.33TGTVATNDD217 pKa = 3.75TLSGDD222 pKa = 3.79GGNTFAVSGNPAHH235 pKa = 6.48GTVTMNADD243 pKa = 3.29GTFTYY248 pKa = 10.36TPAADD253 pKa = 3.69YY254 pKa = 11.27NGADD258 pKa = 3.29SFTYY262 pKa = 10.12TITDD266 pKa = 3.42ADD268 pKa = 4.5GDD270 pKa = 4.11TSTATVNLTVSSVNDD285 pKa = 3.27VPVAVTDD292 pKa = 4.25SINVSEE298 pKa = 4.59EE299 pKa = 3.69GLTNGIGDD307 pKa = 4.16TNGTPTDD314 pKa = 3.77TTNSNSVVGQLTIHH328 pKa = 6.31NPDD331 pKa = 3.33NDD333 pKa = 3.93ALTITLNAPTDD344 pKa = 3.87SLSSGNEE351 pKa = 3.89TIVWSGNNTHH361 pKa = 6.2EE362 pKa = 5.18LIGKK366 pKa = 9.57VGDD369 pKa = 3.7TEE371 pKa = 4.4VIKK374 pKa = 10.68IAIADD379 pKa = 4.0DD380 pKa = 3.82GSYY383 pKa = 10.99SVTLLDD389 pKa = 5.11PVDD392 pKa = 4.31HH393 pKa = 7.16PMNDD397 pKa = 3.2VEE399 pKa = 6.34DD400 pKa = 3.68ILSFNVGVNISDD412 pKa = 4.29GEE414 pKa = 4.38SSSSSTITVNIEE426 pKa = 3.58DD427 pKa = 4.71DD428 pKa = 3.9MPTVEE433 pKa = 4.48TSTTVWTEE441 pKa = 3.5ATSIPDD447 pKa = 3.05IFTGQVSFAGSSANQSRR464 pKa = 11.84FVFANGAVLVTGKK477 pKa = 10.7GFTSDD482 pKa = 3.35TDD484 pKa = 3.63LTLIDD489 pKa = 4.77ADD491 pKa = 4.72LNQSSGGLGVASDD504 pKa = 3.78EE505 pKa = 4.65SPYY508 pKa = 11.0HH509 pKa = 5.72NLANEE514 pKa = 3.64VDD516 pKa = 4.14FRR518 pKa = 11.84KK519 pKa = 7.99TADD522 pKa = 3.5GEE524 pKa = 4.34EE525 pKa = 4.52GSEE528 pKa = 3.84EE529 pKa = 4.91LIIKK533 pKa = 9.88LADD536 pKa = 3.48GKK538 pKa = 10.46ISYY541 pKa = 9.65GATITFDD548 pKa = 3.4YY549 pKa = 10.37MFGGEE554 pKa = 4.28EE555 pKa = 4.09EE556 pKa = 4.12VGEE559 pKa = 4.03ALFYY563 pKa = 10.72RR564 pKa = 11.84DD565 pKa = 3.57GVLISTQRR573 pKa = 11.84FTSDD577 pKa = 3.2AQSGDD582 pKa = 3.33YY583 pKa = 10.86AKK585 pKa = 10.82NFEE588 pKa = 4.34VTDD591 pKa = 3.84GGFDD595 pKa = 3.8TIVIRR600 pKa = 11.84ALDD603 pKa = 3.95NGNGYY608 pKa = 10.01HH609 pKa = 7.43DD610 pKa = 4.98GDD612 pKa = 3.64NSDD615 pKa = 4.09FVVSGVEE622 pKa = 4.04FLGSSQEE629 pKa = 4.04NPISYY634 pKa = 11.0AEE636 pKa = 3.97GTIDD640 pKa = 4.3YY641 pKa = 10.38EE642 pKa = 4.65FGADD646 pKa = 3.45GAGSIVLTSLQNSVRR661 pKa = 11.84LADD664 pKa = 3.6GTEE667 pKa = 3.92PEE669 pKa = 4.38ITISDD674 pKa = 3.57NTIIAKK680 pKa = 10.13DD681 pKa = 3.52ADD683 pKa = 3.56GHH685 pKa = 6.73LVFQIQFTPSTGQWEE700 pKa = 5.18FYY702 pKa = 10.27QYY704 pKa = 11.31QKK706 pKa = 10.32FTIGDD711 pKa = 3.75GSSQTLGFNFRR722 pKa = 11.84VTDD725 pKa = 3.81ADD727 pKa = 3.5GDD729 pKa = 4.32AVNGTVSVGITNHH742 pKa = 4.92GVEE745 pKa = 4.29IEE747 pKa = 4.7GIGAAQGDD755 pKa = 4.36LTVDD759 pKa = 3.56EE760 pKa = 5.51ANLDD764 pKa = 4.38DD765 pKa = 4.06GTQPNEE771 pKa = 3.82SDD773 pKa = 3.39LTQSGTFTLNAMDD786 pKa = 6.08GISEE790 pKa = 4.06IQIGSEE796 pKa = 3.75TFTYY800 pKa = 10.73EE801 pKa = 3.97EE802 pKa = 4.54LVNAASNNISIEE814 pKa = 4.13TDD816 pKa = 2.69HH817 pKa = 6.48GTLVITGYY825 pKa = 10.32SGSEE829 pKa = 3.76TSGTVSYY836 pKa = 10.93SYY838 pKa = 9.46TLKK841 pKa = 11.05DD842 pKa = 3.48SVDD845 pKa = 3.61NDD847 pKa = 3.97TQNGATSDD855 pKa = 3.93EE856 pKa = 4.51YY857 pKa = 11.72VDD859 pKa = 4.01QIDD862 pKa = 4.55VNVTDD867 pKa = 4.59KK868 pKa = 11.64DD869 pKa = 3.84GDD871 pKa = 3.9SSSASININIQDD883 pKa = 4.34DD884 pKa = 4.21APVASDD890 pKa = 3.24TTVSVDD896 pKa = 3.21AMQEE900 pKa = 4.23TVFNANVMLTLDD912 pKa = 3.82YY913 pKa = 10.47SGSMSSTALSEE924 pKa = 4.29QKK926 pKa = 10.58DD927 pKa = 3.65AAKK930 pKa = 10.6KK931 pKa = 10.78LLDD934 pKa = 3.67QYY936 pKa = 11.39QKK938 pKa = 11.22SLNEE942 pKa = 3.85ADD944 pKa = 3.88RR945 pKa = 11.84GDD947 pKa = 3.81VKK949 pKa = 11.64VMVIKK954 pKa = 10.39FATGAATHH962 pKa = 5.91EE963 pKa = 4.64KK964 pKa = 9.85VWLTIAEE971 pKa = 4.08AKK973 pKa = 10.47AYY975 pKa = 9.41IDD977 pKa = 3.04QGKK980 pKa = 9.14PYY982 pKa = 10.55GIDD985 pKa = 2.9SGTNYY990 pKa = 10.88DD991 pKa = 4.43DD992 pKa = 4.08ALKK995 pKa = 9.65TLIEE999 pKa = 4.63NYY1001 pKa = 9.95SSSGKK1006 pKa = 8.38VTAEE1010 pKa = 3.59GTINNSYY1017 pKa = 10.82FMTDD1021 pKa = 3.47GQPTQSNYY1029 pKa = 9.53TSSNDD1034 pKa = 3.25GSKK1037 pKa = 9.38TEE1039 pKa = 4.29DD1040 pKa = 3.41SKK1042 pKa = 12.01GDD1044 pKa = 3.64GIGIGEE1050 pKa = 4.46HH1051 pKa = 6.58SNNPYY1056 pKa = 9.53YY1057 pKa = 10.94AQGTDD1062 pKa = 3.56ANSTTYY1068 pKa = 10.81VGTNGEE1074 pKa = 4.21VGQGNWQAFLEE1085 pKa = 4.2EE1086 pKa = 4.46HH1087 pKa = 7.35DD1088 pKa = 3.77IVSYY1092 pKa = 10.57AIGMGNNVQSSGLEE1106 pKa = 3.89PIAYY1110 pKa = 9.58DD1111 pKa = 3.59GTSEE1115 pKa = 4.22QQNHH1119 pKa = 6.82DD1120 pKa = 4.48KK1121 pKa = 10.94LLQPQVDD1128 pKa = 4.44FNKK1131 pKa = 10.79LGDD1134 pKa = 4.3LLVSTTPEE1142 pKa = 4.05AQPVTSTLSAEE1153 pKa = 4.13LGADD1157 pKa = 3.31GGYY1160 pKa = 10.19VSSITISGEE1169 pKa = 3.86TYY1171 pKa = 10.05TFDD1174 pKa = 3.26GTTITNPNGTTANGHH1189 pKa = 4.99TLCVTTEE1196 pKa = 4.13FGTFEE1201 pKa = 4.39INLEE1205 pKa = 4.28TGVYY1209 pKa = 9.55TYY1211 pKa = 10.96TPVEE1215 pKa = 3.85ALAQVEE1221 pKa = 4.23QEE1223 pKa = 4.44TIGFTVTDD1231 pKa = 3.69NDD1233 pKa = 4.26GDD1235 pKa = 4.04TSSATLTIDD1244 pKa = 4.62LSALVPDD1251 pKa = 4.21VPTVVEE1257 pKa = 4.48DD1258 pKa = 3.45AHH1260 pKa = 6.44DD1261 pKa = 3.67TVITNEE1267 pKa = 4.11NGASSIQAEE1276 pKa = 4.12ASLGVVVGKK1285 pKa = 10.35DD1286 pKa = 3.36SAGSTVTVTNANGDD1300 pKa = 4.09SLVGDD1305 pKa = 4.07TVTVKK1310 pKa = 9.33VTDD1313 pKa = 4.02ANGSEE1318 pKa = 4.26HH1319 pKa = 7.28EE1320 pKa = 4.6INATYY1325 pKa = 11.25NDD1327 pKa = 3.48VQLKK1331 pKa = 9.78YY1332 pKa = 10.61ISDD1335 pKa = 3.78GSGGLIAVTDD1345 pKa = 4.49DD1346 pKa = 3.06ATQTHH1351 pKa = 6.08VFTVEE1356 pKa = 3.64GDD1358 pKa = 3.61ASAGTYY1364 pKa = 9.32KK1365 pKa = 9.56VTMIHH1370 pKa = 6.8ALDD1373 pKa = 3.91PVYY1376 pKa = 10.56SVPSILEE1383 pKa = 4.1DD1384 pKa = 3.12QTTQISLNSNGGSNSGINVTLNSNGGAVNQYY1415 pKa = 10.45GNSIGINDD1423 pKa = 4.16YY1424 pKa = 11.32DD1425 pKa = 3.84VGSAKK1430 pKa = 10.75YY1431 pKa = 9.45EE1432 pKa = 3.84IEE1434 pKa = 4.42GYY1436 pKa = 10.03EE1437 pKa = 4.05VLSMNFNALTGVSVSKK1453 pKa = 11.0VSVNFNDD1460 pKa = 4.59FGYY1463 pKa = 10.79HH1464 pKa = 6.91DD1465 pKa = 4.17DD1466 pKa = 5.23AIVKK1470 pKa = 9.41VYY1472 pKa = 9.47YY1473 pKa = 10.1TDD1475 pKa = 3.43NTYY1478 pKa = 9.23DD1479 pKa = 3.19TYY1481 pKa = 11.27EE1482 pKa = 3.72VHH1484 pKa = 7.28GDD1486 pKa = 3.36YY1487 pKa = 10.88NDD1489 pKa = 3.89RR1490 pKa = 11.84DD1491 pKa = 4.26GIDD1494 pKa = 3.08IEE1496 pKa = 4.24IPEE1499 pKa = 4.79GKK1501 pKa = 7.78TLSRR1505 pKa = 11.84IDD1507 pKa = 4.52FGAEE1511 pKa = 3.79DD1512 pKa = 4.7YY1513 pKa = 11.65YY1514 pKa = 11.59DD1515 pKa = 4.55DD1516 pKa = 4.54YY1517 pKa = 11.83SVNSTITVVYY1527 pKa = 9.62DD1528 pKa = 3.45KK1529 pKa = 11.22QSTIPVDD1536 pKa = 3.61NPEE1539 pKa = 3.84QTLEE1543 pKa = 4.19FGAVVTDD1550 pKa = 3.93GNGDD1554 pKa = 3.24QSEE1557 pKa = 4.69VVTFDD1562 pKa = 3.12VTVDD1566 pKa = 3.5SDD1568 pKa = 3.9HH1569 pKa = 6.65VLQGDD1574 pKa = 3.75VSDD1577 pKa = 5.17SIIGGDD1583 pKa = 3.77GDD1585 pKa = 4.63DD1586 pKa = 3.99TFKK1589 pKa = 11.37LLNGQDD1595 pKa = 3.36INFDD1599 pKa = 3.61GLDD1602 pKa = 3.35AHH1604 pKa = 7.3IKK1606 pKa = 10.21NIEE1609 pKa = 4.23TIDD1612 pKa = 3.77LSAEE1616 pKa = 3.97GKK1618 pKa = 10.23NEE1620 pKa = 3.74IKK1622 pKa = 10.62NLSLDD1627 pKa = 3.92DD1628 pKa = 4.76VIDD1631 pKa = 3.66MTDD1634 pKa = 3.0SDD1636 pKa = 4.55NEE1638 pKa = 4.05IKK1640 pKa = 10.48ISGTSEE1646 pKa = 4.66DD1647 pKa = 4.24EE1648 pKa = 4.11VHH1650 pKa = 6.52LTNEE1654 pKa = 3.44WSTNNVVDD1662 pKa = 3.59ADD1664 pKa = 4.4GYY1666 pKa = 10.49IEE1668 pKa = 4.8YY1669 pKa = 10.06IGTSDD1674 pKa = 4.71DD1675 pKa = 3.36EE1676 pKa = 4.59TVKK1679 pKa = 11.01VKK1681 pKa = 10.1IHH1683 pKa = 5.93QDD1685 pKa = 2.39IHH1687 pKa = 7.2TDD1689 pKa = 3.08IQQ1691 pKa = 3.44
MM1 pKa = 7.67AEE3 pKa = 4.54SIAKK7 pKa = 8.93VQSLLGKK14 pKa = 9.84FYY16 pKa = 11.11AQDD19 pKa = 2.98EE20 pKa = 4.73SGNVRR25 pKa = 11.84VLKK28 pKa = 11.02VGDD31 pKa = 3.75TIYY34 pKa = 10.57QGEE37 pKa = 4.57KK38 pKa = 10.47VYY40 pKa = 11.35GDD42 pKa = 3.7AANNSSDD49 pKa = 3.21KK50 pKa = 11.09LQIVSDD56 pKa = 3.83TQEE59 pKa = 4.0TFDD62 pKa = 3.63ITGSDD67 pKa = 3.36SLVFDD72 pKa = 4.23TSTNEE77 pKa = 3.58ATFGNEE83 pKa = 2.93EE84 pKa = 4.07VAFNPNSAWMPTADD98 pKa = 3.42NATVPEE104 pKa = 4.79GEE106 pKa = 4.68TVTDD110 pKa = 3.71AQGDD114 pKa = 3.65ITEE117 pKa = 4.65EE118 pKa = 3.63EE119 pKa = 4.11TAAGNEE125 pKa = 4.17AASSSDD131 pKa = 2.99VGVFNFEE138 pKa = 3.97NRR140 pKa = 11.84DD141 pKa = 3.35GDD143 pKa = 4.25SVNIQSDD150 pKa = 3.27LRR152 pKa = 11.84EE153 pKa = 4.19SVFSNNSDD161 pKa = 3.28SLVSNNGDD169 pKa = 3.43GLNSNPSSNLLLNGTPTPDD188 pKa = 3.65DD189 pKa = 4.12VNDD192 pKa = 3.52VPVAVDD198 pKa = 4.61DD199 pKa = 5.16SFTLDD204 pKa = 3.21EE205 pKa = 4.74DD206 pKa = 3.78ASYY209 pKa = 10.33TGTVATNDD217 pKa = 3.75TLSGDD222 pKa = 3.79GGNTFAVSGNPAHH235 pKa = 6.48GTVTMNADD243 pKa = 3.29GTFTYY248 pKa = 10.36TPAADD253 pKa = 3.69YY254 pKa = 11.27NGADD258 pKa = 3.29SFTYY262 pKa = 10.12TITDD266 pKa = 3.42ADD268 pKa = 4.5GDD270 pKa = 4.11TSTATVNLTVSSVNDD285 pKa = 3.27VPVAVTDD292 pKa = 4.25SINVSEE298 pKa = 4.59EE299 pKa = 3.69GLTNGIGDD307 pKa = 4.16TNGTPTDD314 pKa = 3.77TTNSNSVVGQLTIHH328 pKa = 6.31NPDD331 pKa = 3.33NDD333 pKa = 3.93ALTITLNAPTDD344 pKa = 3.87SLSSGNEE351 pKa = 3.89TIVWSGNNTHH361 pKa = 6.2EE362 pKa = 5.18LIGKK366 pKa = 9.57VGDD369 pKa = 3.7TEE371 pKa = 4.4VIKK374 pKa = 10.68IAIADD379 pKa = 4.0DD380 pKa = 3.82GSYY383 pKa = 10.99SVTLLDD389 pKa = 5.11PVDD392 pKa = 4.31HH393 pKa = 7.16PMNDD397 pKa = 3.2VEE399 pKa = 6.34DD400 pKa = 3.68ILSFNVGVNISDD412 pKa = 4.29GEE414 pKa = 4.38SSSSSTITVNIEE426 pKa = 3.58DD427 pKa = 4.71DD428 pKa = 3.9MPTVEE433 pKa = 4.48TSTTVWTEE441 pKa = 3.5ATSIPDD447 pKa = 3.05IFTGQVSFAGSSANQSRR464 pKa = 11.84FVFANGAVLVTGKK477 pKa = 10.7GFTSDD482 pKa = 3.35TDD484 pKa = 3.63LTLIDD489 pKa = 4.77ADD491 pKa = 4.72LNQSSGGLGVASDD504 pKa = 3.78EE505 pKa = 4.65SPYY508 pKa = 11.0HH509 pKa = 5.72NLANEE514 pKa = 3.64VDD516 pKa = 4.14FRR518 pKa = 11.84KK519 pKa = 7.99TADD522 pKa = 3.5GEE524 pKa = 4.34EE525 pKa = 4.52GSEE528 pKa = 3.84EE529 pKa = 4.91LIIKK533 pKa = 9.88LADD536 pKa = 3.48GKK538 pKa = 10.46ISYY541 pKa = 9.65GATITFDD548 pKa = 3.4YY549 pKa = 10.37MFGGEE554 pKa = 4.28EE555 pKa = 4.09EE556 pKa = 4.12VGEE559 pKa = 4.03ALFYY563 pKa = 10.72RR564 pKa = 11.84DD565 pKa = 3.57GVLISTQRR573 pKa = 11.84FTSDD577 pKa = 3.2AQSGDD582 pKa = 3.33YY583 pKa = 10.86AKK585 pKa = 10.82NFEE588 pKa = 4.34VTDD591 pKa = 3.84GGFDD595 pKa = 3.8TIVIRR600 pKa = 11.84ALDD603 pKa = 3.95NGNGYY608 pKa = 10.01HH609 pKa = 7.43DD610 pKa = 4.98GDD612 pKa = 3.64NSDD615 pKa = 4.09FVVSGVEE622 pKa = 4.04FLGSSQEE629 pKa = 4.04NPISYY634 pKa = 11.0AEE636 pKa = 3.97GTIDD640 pKa = 4.3YY641 pKa = 10.38EE642 pKa = 4.65FGADD646 pKa = 3.45GAGSIVLTSLQNSVRR661 pKa = 11.84LADD664 pKa = 3.6GTEE667 pKa = 3.92PEE669 pKa = 4.38ITISDD674 pKa = 3.57NTIIAKK680 pKa = 10.13DD681 pKa = 3.52ADD683 pKa = 3.56GHH685 pKa = 6.73LVFQIQFTPSTGQWEE700 pKa = 5.18FYY702 pKa = 10.27QYY704 pKa = 11.31QKK706 pKa = 10.32FTIGDD711 pKa = 3.75GSSQTLGFNFRR722 pKa = 11.84VTDD725 pKa = 3.81ADD727 pKa = 3.5GDD729 pKa = 4.32AVNGTVSVGITNHH742 pKa = 4.92GVEE745 pKa = 4.29IEE747 pKa = 4.7GIGAAQGDD755 pKa = 4.36LTVDD759 pKa = 3.56EE760 pKa = 5.51ANLDD764 pKa = 4.38DD765 pKa = 4.06GTQPNEE771 pKa = 3.82SDD773 pKa = 3.39LTQSGTFTLNAMDD786 pKa = 6.08GISEE790 pKa = 4.06IQIGSEE796 pKa = 3.75TFTYY800 pKa = 10.73EE801 pKa = 3.97EE802 pKa = 4.54LVNAASNNISIEE814 pKa = 4.13TDD816 pKa = 2.69HH817 pKa = 6.48GTLVITGYY825 pKa = 10.32SGSEE829 pKa = 3.76TSGTVSYY836 pKa = 10.93SYY838 pKa = 9.46TLKK841 pKa = 11.05DD842 pKa = 3.48SVDD845 pKa = 3.61NDD847 pKa = 3.97TQNGATSDD855 pKa = 3.93EE856 pKa = 4.51YY857 pKa = 11.72VDD859 pKa = 4.01QIDD862 pKa = 4.55VNVTDD867 pKa = 4.59KK868 pKa = 11.64DD869 pKa = 3.84GDD871 pKa = 3.9SSSASININIQDD883 pKa = 4.34DD884 pKa = 4.21APVASDD890 pKa = 3.24TTVSVDD896 pKa = 3.21AMQEE900 pKa = 4.23TVFNANVMLTLDD912 pKa = 3.82YY913 pKa = 10.47SGSMSSTALSEE924 pKa = 4.29QKK926 pKa = 10.58DD927 pKa = 3.65AAKK930 pKa = 10.6KK931 pKa = 10.78LLDD934 pKa = 3.67QYY936 pKa = 11.39QKK938 pKa = 11.22SLNEE942 pKa = 3.85ADD944 pKa = 3.88RR945 pKa = 11.84GDD947 pKa = 3.81VKK949 pKa = 11.64VMVIKK954 pKa = 10.39FATGAATHH962 pKa = 5.91EE963 pKa = 4.64KK964 pKa = 9.85VWLTIAEE971 pKa = 4.08AKK973 pKa = 10.47AYY975 pKa = 9.41IDD977 pKa = 3.04QGKK980 pKa = 9.14PYY982 pKa = 10.55GIDD985 pKa = 2.9SGTNYY990 pKa = 10.88DD991 pKa = 4.43DD992 pKa = 4.08ALKK995 pKa = 9.65TLIEE999 pKa = 4.63NYY1001 pKa = 9.95SSSGKK1006 pKa = 8.38VTAEE1010 pKa = 3.59GTINNSYY1017 pKa = 10.82FMTDD1021 pKa = 3.47GQPTQSNYY1029 pKa = 9.53TSSNDD1034 pKa = 3.25GSKK1037 pKa = 9.38TEE1039 pKa = 4.29DD1040 pKa = 3.41SKK1042 pKa = 12.01GDD1044 pKa = 3.64GIGIGEE1050 pKa = 4.46HH1051 pKa = 6.58SNNPYY1056 pKa = 9.53YY1057 pKa = 10.94AQGTDD1062 pKa = 3.56ANSTTYY1068 pKa = 10.81VGTNGEE1074 pKa = 4.21VGQGNWQAFLEE1085 pKa = 4.2EE1086 pKa = 4.46HH1087 pKa = 7.35DD1088 pKa = 3.77IVSYY1092 pKa = 10.57AIGMGNNVQSSGLEE1106 pKa = 3.89PIAYY1110 pKa = 9.58DD1111 pKa = 3.59GTSEE1115 pKa = 4.22QQNHH1119 pKa = 6.82DD1120 pKa = 4.48KK1121 pKa = 10.94LLQPQVDD1128 pKa = 4.44FNKK1131 pKa = 10.79LGDD1134 pKa = 4.3LLVSTTPEE1142 pKa = 4.05AQPVTSTLSAEE1153 pKa = 4.13LGADD1157 pKa = 3.31GGYY1160 pKa = 10.19VSSITISGEE1169 pKa = 3.86TYY1171 pKa = 10.05TFDD1174 pKa = 3.26GTTITNPNGTTANGHH1189 pKa = 4.99TLCVTTEE1196 pKa = 4.13FGTFEE1201 pKa = 4.39INLEE1205 pKa = 4.28TGVYY1209 pKa = 9.55TYY1211 pKa = 10.96TPVEE1215 pKa = 3.85ALAQVEE1221 pKa = 4.23QEE1223 pKa = 4.44TIGFTVTDD1231 pKa = 3.69NDD1233 pKa = 4.26GDD1235 pKa = 4.04TSSATLTIDD1244 pKa = 4.62LSALVPDD1251 pKa = 4.21VPTVVEE1257 pKa = 4.48DD1258 pKa = 3.45AHH1260 pKa = 6.44DD1261 pKa = 3.67TVITNEE1267 pKa = 4.11NGASSIQAEE1276 pKa = 4.12ASLGVVVGKK1285 pKa = 10.35DD1286 pKa = 3.36SAGSTVTVTNANGDD1300 pKa = 4.09SLVGDD1305 pKa = 4.07TVTVKK1310 pKa = 9.33VTDD1313 pKa = 4.02ANGSEE1318 pKa = 4.26HH1319 pKa = 7.28EE1320 pKa = 4.6INATYY1325 pKa = 11.25NDD1327 pKa = 3.48VQLKK1331 pKa = 9.78YY1332 pKa = 10.61ISDD1335 pKa = 3.78GSGGLIAVTDD1345 pKa = 4.49DD1346 pKa = 3.06ATQTHH1351 pKa = 6.08VFTVEE1356 pKa = 3.64GDD1358 pKa = 3.61ASAGTYY1364 pKa = 9.32KK1365 pKa = 9.56VTMIHH1370 pKa = 6.8ALDD1373 pKa = 3.91PVYY1376 pKa = 10.56SVPSILEE1383 pKa = 4.1DD1384 pKa = 3.12QTTQISLNSNGGSNSGINVTLNSNGGAVNQYY1415 pKa = 10.45GNSIGINDD1423 pKa = 4.16YY1424 pKa = 11.32DD1425 pKa = 3.84VGSAKK1430 pKa = 10.75YY1431 pKa = 9.45EE1432 pKa = 3.84IEE1434 pKa = 4.42GYY1436 pKa = 10.03EE1437 pKa = 4.05VLSMNFNALTGVSVSKK1453 pKa = 11.0VSVNFNDD1460 pKa = 4.59FGYY1463 pKa = 10.79HH1464 pKa = 6.91DD1465 pKa = 4.17DD1466 pKa = 5.23AIVKK1470 pKa = 9.41VYY1472 pKa = 9.47YY1473 pKa = 10.1TDD1475 pKa = 3.43NTYY1478 pKa = 9.23DD1479 pKa = 3.19TYY1481 pKa = 11.27EE1482 pKa = 3.72VHH1484 pKa = 7.28GDD1486 pKa = 3.36YY1487 pKa = 10.88NDD1489 pKa = 3.89RR1490 pKa = 11.84DD1491 pKa = 4.26GIDD1494 pKa = 3.08IEE1496 pKa = 4.24IPEE1499 pKa = 4.79GKK1501 pKa = 7.78TLSRR1505 pKa = 11.84IDD1507 pKa = 4.52FGAEE1511 pKa = 3.79DD1512 pKa = 4.7YY1513 pKa = 11.65YY1514 pKa = 11.59DD1515 pKa = 4.55DD1516 pKa = 4.54YY1517 pKa = 11.83SVNSTITVVYY1527 pKa = 9.62DD1528 pKa = 3.45KK1529 pKa = 11.22QSTIPVDD1536 pKa = 3.61NPEE1539 pKa = 3.84QTLEE1543 pKa = 4.19FGAVVTDD1550 pKa = 3.93GNGDD1554 pKa = 3.24QSEE1557 pKa = 4.69VVTFDD1562 pKa = 3.12VTVDD1566 pKa = 3.5SDD1568 pKa = 3.9HH1569 pKa = 6.65VLQGDD1574 pKa = 3.75VSDD1577 pKa = 5.17SIIGGDD1583 pKa = 3.77GDD1585 pKa = 4.63DD1586 pKa = 3.99TFKK1589 pKa = 11.37LLNGQDD1595 pKa = 3.36INFDD1599 pKa = 3.61GLDD1602 pKa = 3.35AHH1604 pKa = 7.3IKK1606 pKa = 10.21NIEE1609 pKa = 4.23TIDD1612 pKa = 3.77LSAEE1616 pKa = 3.97GKK1618 pKa = 10.23NEE1620 pKa = 3.74IKK1622 pKa = 10.62NLSLDD1627 pKa = 3.92DD1628 pKa = 4.76VIDD1631 pKa = 3.66MTDD1634 pKa = 3.0SDD1636 pKa = 4.55NEE1638 pKa = 4.05IKK1640 pKa = 10.48ISGTSEE1646 pKa = 4.66DD1647 pKa = 4.24EE1648 pKa = 4.11VHH1650 pKa = 6.52LTNEE1654 pKa = 3.44WSTNNVVDD1662 pKa = 3.59ADD1664 pKa = 4.4GYY1666 pKa = 10.49IEE1668 pKa = 4.8YY1669 pKa = 10.06IGTSDD1674 pKa = 4.71DD1675 pKa = 3.36EE1676 pKa = 4.59TVKK1679 pKa = 11.01VKK1681 pKa = 10.1IHH1683 pKa = 5.93QDD1685 pKa = 2.39IHH1687 pKa = 7.2TDD1689 pKa = 3.08IQQ1691 pKa = 3.44
Molecular weight: 178.88 kDa
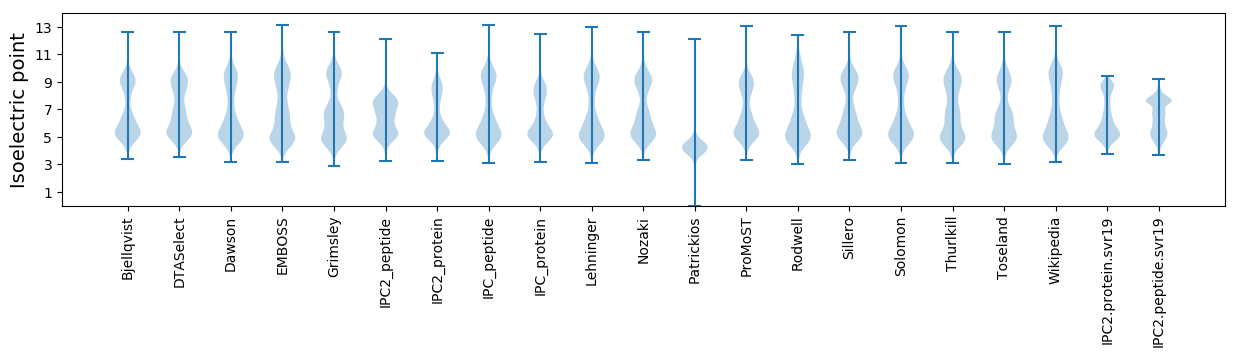
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q0XQW7|A0A4Q0XQW7_9PROT Bcr/CflA family efflux transporter OS=Arcobacter sp. CECT 8987 OX=2044504 GN=CRV04_06635 PE=3 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.31RR14 pKa = 11.84THH16 pKa = 5.88GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.73TKK25 pKa = 10.05SGRR28 pKa = 11.84KK29 pKa = 8.71VISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.87GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.31RR14 pKa = 11.84THH16 pKa = 5.88GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.73TKK25 pKa = 10.05SGRR28 pKa = 11.84KK29 pKa = 8.71VISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.87GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
787083 |
37 |
1691 |
316.6 |
35.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.481 ± 0.062 | 0.962 ± 0.018 |
5.292 ± 0.04 | 7.266 ± 0.047 |
5.161 ± 0.044 | 5.513 ± 0.06 |
2.049 ± 0.024 | 8.576 ± 0.052 |
8.449 ± 0.054 | 9.825 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.568 ± 0.024 | 5.663 ± 0.039 |
2.896 ± 0.029 | 3.695 ± 0.033 |
3.033 ± 0.028 | 6.28 ± 0.033 |
5.298 ± 0.042 | 6.268 ± 0.035 |
0.724 ± 0.017 | 4.002 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
