
Kutzneria sp. 744
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Kutzneria; unclassified Kutzneria
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
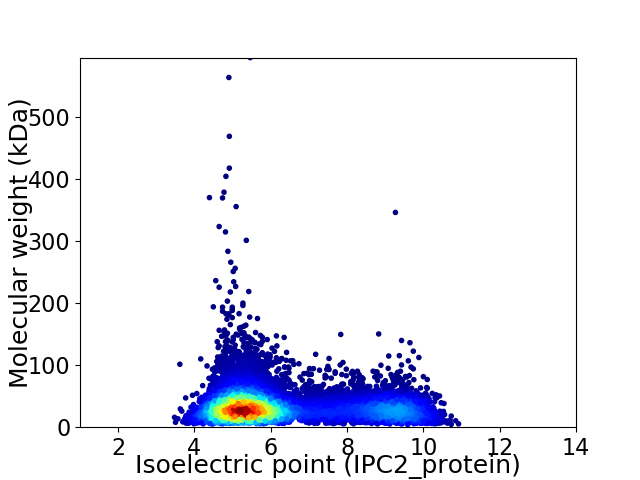
Virtual 2D-PAGE plot for 10148 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
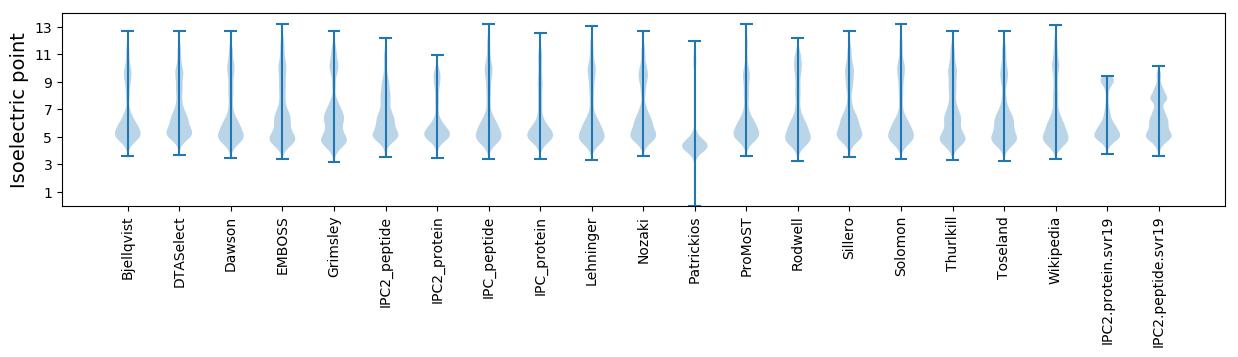
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W7T443|W7T443_9PSEU Uncharacterized protein OS=Kutzneria sp. 744 OX=345341 GN=KUTG_08397 PE=4 SV=1
MM1 pKa = 7.27ATGISLLDD9 pKa = 4.31FIMEE13 pKa = 4.51LLGDD17 pKa = 3.76QSARR21 pKa = 11.84DD22 pKa = 3.75HH23 pKa = 6.9FEE25 pKa = 4.55SNPQQVLNDD34 pKa = 3.47HH35 pKa = 6.59GFNHH39 pKa = 6.86LCAADD44 pKa = 3.55VHH46 pKa = 6.81DD47 pKa = 5.69AMPLVIDD54 pKa = 4.61AAQSFDD60 pKa = 3.14RR61 pKa = 11.84TYY63 pKa = 11.3DD64 pKa = 3.02AGGPSHH70 pKa = 7.02VVTPPPPPPVHH81 pKa = 6.53GGGGMEE87 pKa = 4.5HH88 pKa = 7.49AIEE91 pKa = 4.15QIRR94 pKa = 11.84YY95 pKa = 6.37ITQNYY100 pKa = 8.7SYY102 pKa = 11.2SVDD105 pKa = 3.12SHH107 pKa = 5.48NTVVDD112 pKa = 3.33NSVNQNVWARR122 pKa = 11.84DD123 pKa = 3.53VFQSFDD129 pKa = 3.2NHH131 pKa = 6.09SVVTTGDD138 pKa = 3.52HH139 pKa = 5.68NVVGNGNEE147 pKa = 4.12VQNGDD152 pKa = 3.4LTADD156 pKa = 3.11HH157 pKa = 6.9GGAVAVGGSATGSNDD172 pKa = 3.43DD173 pKa = 3.9NSTDD177 pKa = 3.1VHH179 pKa = 7.59AYY181 pKa = 10.31GSGDD185 pKa = 3.25VATATNGSSLSQTDD199 pKa = 3.2SHH201 pKa = 6.26NTTDD205 pKa = 3.29SHH207 pKa = 7.33NSTDD211 pKa = 3.41SHH213 pKa = 6.81NDD215 pKa = 2.76SHH217 pKa = 7.48NNTAVGSGNDD227 pKa = 3.38SHH229 pKa = 7.57DD230 pKa = 3.58NTAVGSGNDD239 pKa = 3.37SHH241 pKa = 7.56DD242 pKa = 3.39NTAIGSGNDD251 pKa = 3.32SHH253 pKa = 7.9DD254 pKa = 4.28DD255 pKa = 3.54SHH257 pKa = 8.23DD258 pKa = 3.57NTAVGSGNDD267 pKa = 3.38SHH269 pKa = 7.58DD270 pKa = 3.57NTAVDD275 pKa = 4.35SGNDD279 pKa = 3.42SHH281 pKa = 7.78DD282 pKa = 4.37DD283 pKa = 3.41SHH285 pKa = 9.28DD286 pKa = 3.88DD287 pKa = 3.38VNTAIGSGNDD297 pKa = 3.17SHH299 pKa = 9.27DD300 pKa = 5.94DD301 pKa = 3.29IASDD305 pKa = 3.55NNTAIGSGNDD315 pKa = 3.34SHH317 pKa = 7.18DD318 pKa = 3.7HH319 pKa = 6.34SLTEE323 pKa = 4.44LDD325 pKa = 3.7SHH327 pKa = 7.1NDD329 pKa = 2.86TDD331 pKa = 3.88VASNNDD337 pKa = 2.88IASNNEE343 pKa = 3.23VDD345 pKa = 4.27HH346 pKa = 6.46NLIHH350 pKa = 7.02II351 pKa = 4.72
MM1 pKa = 7.27ATGISLLDD9 pKa = 4.31FIMEE13 pKa = 4.51LLGDD17 pKa = 3.76QSARR21 pKa = 11.84DD22 pKa = 3.75HH23 pKa = 6.9FEE25 pKa = 4.55SNPQQVLNDD34 pKa = 3.47HH35 pKa = 6.59GFNHH39 pKa = 6.86LCAADD44 pKa = 3.55VHH46 pKa = 6.81DD47 pKa = 5.69AMPLVIDD54 pKa = 4.61AAQSFDD60 pKa = 3.14RR61 pKa = 11.84TYY63 pKa = 11.3DD64 pKa = 3.02AGGPSHH70 pKa = 7.02VVTPPPPPPVHH81 pKa = 6.53GGGGMEE87 pKa = 4.5HH88 pKa = 7.49AIEE91 pKa = 4.15QIRR94 pKa = 11.84YY95 pKa = 6.37ITQNYY100 pKa = 8.7SYY102 pKa = 11.2SVDD105 pKa = 3.12SHH107 pKa = 5.48NTVVDD112 pKa = 3.33NSVNQNVWARR122 pKa = 11.84DD123 pKa = 3.53VFQSFDD129 pKa = 3.2NHH131 pKa = 6.09SVVTTGDD138 pKa = 3.52HH139 pKa = 5.68NVVGNGNEE147 pKa = 4.12VQNGDD152 pKa = 3.4LTADD156 pKa = 3.11HH157 pKa = 6.9GGAVAVGGSATGSNDD172 pKa = 3.43DD173 pKa = 3.9NSTDD177 pKa = 3.1VHH179 pKa = 7.59AYY181 pKa = 10.31GSGDD185 pKa = 3.25VATATNGSSLSQTDD199 pKa = 3.2SHH201 pKa = 6.26NTTDD205 pKa = 3.29SHH207 pKa = 7.33NSTDD211 pKa = 3.41SHH213 pKa = 6.81NDD215 pKa = 2.76SHH217 pKa = 7.48NNTAVGSGNDD227 pKa = 3.38SHH229 pKa = 7.57DD230 pKa = 3.58NTAVGSGNDD239 pKa = 3.37SHH241 pKa = 7.56DD242 pKa = 3.39NTAIGSGNDD251 pKa = 3.32SHH253 pKa = 7.9DD254 pKa = 4.28DD255 pKa = 3.54SHH257 pKa = 8.23DD258 pKa = 3.57NTAVGSGNDD267 pKa = 3.38SHH269 pKa = 7.58DD270 pKa = 3.57NTAVDD275 pKa = 4.35SGNDD279 pKa = 3.42SHH281 pKa = 7.78DD282 pKa = 4.37DD283 pKa = 3.41SHH285 pKa = 9.28DD286 pKa = 3.88DD287 pKa = 3.38VNTAIGSGNDD297 pKa = 3.17SHH299 pKa = 9.27DD300 pKa = 5.94DD301 pKa = 3.29IASDD305 pKa = 3.55NNTAIGSGNDD315 pKa = 3.34SHH317 pKa = 7.18DD318 pKa = 3.7HH319 pKa = 6.34SLTEE323 pKa = 4.44LDD325 pKa = 3.7SHH327 pKa = 7.1NDD329 pKa = 2.86TDD331 pKa = 3.88VASNNDD337 pKa = 2.88IASNNEE343 pKa = 3.23VDD345 pKa = 4.27HH346 pKa = 6.46NLIHH350 pKa = 7.02II351 pKa = 4.72
Molecular weight: 36.56 kDa
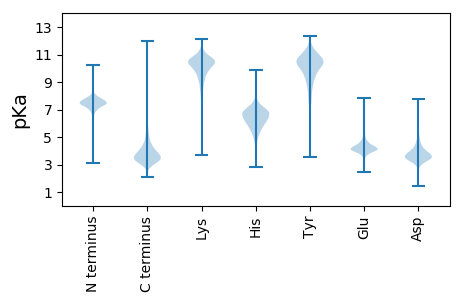
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W7T212|W7T212_9PSEU Uncharacterized protein OS=Kutzneria sp. 744 OX=345341 GN=KUTG_04410 PE=4 SV=1
MM1 pKa = 6.62NTAAAPIQTGTGALGGGSGGGSTGGGTGGGTRR33 pKa = 11.84VAAARR38 pKa = 11.84RR39 pKa = 11.84AGPGAPAVRR48 pKa = 11.84PAAAPRR54 pKa = 11.84AAPARR59 pKa = 11.84AAAA62 pKa = 4.2
MM1 pKa = 6.62NTAAAPIQTGTGALGGGSGGGSTGGGTGGGTRR33 pKa = 11.84VAAARR38 pKa = 11.84RR39 pKa = 11.84AGPGAPAVRR48 pKa = 11.84PAAAPRR54 pKa = 11.84AAPARR59 pKa = 11.84AAAA62 pKa = 4.2
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3322608 |
37 |
5422 |
327.4 |
35.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.896 ± 0.034 | 0.858 ± 0.008 |
6.169 ± 0.02 | 4.964 ± 0.02 |
2.904 ± 0.013 | 9.057 ± 0.027 |
2.356 ± 0.012 | 3.509 ± 0.016 |
2.023 ± 0.015 | 10.359 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.813 ± 0.009 | 2.19 ± 0.017 |
5.809 ± 0.021 | 3.122 ± 0.015 |
7.658 ± 0.031 | 5.432 ± 0.021 |
6.248 ± 0.022 | 8.938 ± 0.026 |
1.621 ± 0.01 | 2.074 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
