
Citromicrobium phage vB_Cib_ssDNA_P1
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
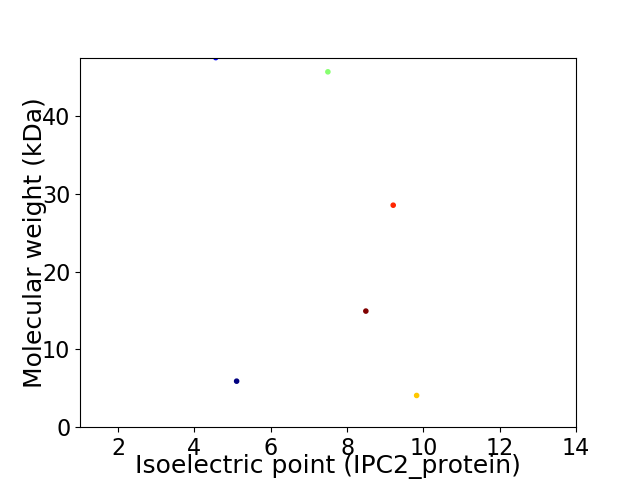
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4PJ67|A0A2H4PJ67_9VIRU Replication initiator OS=Citromicrobium phage vB_Cib_ssDNA_P1 OX=2053684 PE=4 SV=1
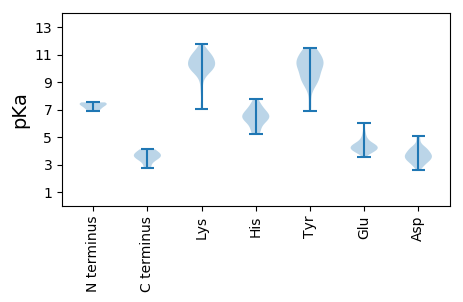
MM1 pKa = 7.29EE2 pKa = 5.9NIMRR6 pKa = 11.84TLMSAPPTGRR16 pKa = 11.84VGRR19 pKa = 11.84YY20 pKa = 6.92PKK22 pKa = 10.55HH23 pKa = 6.4PFLTQEE29 pKa = 4.55KK30 pKa = 9.31PFTAQPFFLARR41 pKa = 11.84VLPGEE46 pKa = 4.11TLQNLRR52 pKa = 11.84LEE54 pKa = 4.5SRR56 pKa = 11.84VITDD60 pKa = 4.82PIKK63 pKa = 10.91NSIIGWSKK71 pKa = 9.69EE72 pKa = 3.73YY73 pKa = 10.58YY74 pKa = 8.92VFYY77 pKa = 11.25VKK79 pKa = 10.76VSDD82 pKa = 5.08LMIDD86 pKa = 4.11AIKK89 pKa = 11.15DD90 pKa = 3.46MFIDD94 pKa = 3.67PTNAEE99 pKa = 3.89ITGQDD104 pKa = 2.85AATNVQRR111 pKa = 11.84TYY113 pKa = 10.3TPQGGIDD120 pKa = 3.54WTGMCLTRR128 pKa = 11.84IVEE131 pKa = 4.41TYY133 pKa = 10.38FRR135 pKa = 11.84DD136 pKa = 3.7EE137 pKa = 4.45GEE139 pKa = 4.33TAASHH144 pKa = 6.52ALADD148 pKa = 4.14GTPMVQIRR156 pKa = 11.84QNSFLDD162 pKa = 4.03SITDD166 pKa = 3.31KK167 pKa = 11.77DD168 pKa = 4.48EE169 pKa = 4.43IPEE172 pKa = 4.17GAAISTATDD181 pKa = 3.13AGDD184 pKa = 4.34LDD186 pKa = 4.13RR187 pKa = 11.84LMDD190 pKa = 4.35AFEE193 pKa = 4.39MARR196 pKa = 11.84SLGMANMSYY205 pKa = 11.15EE206 pKa = 4.15DD207 pKa = 3.71FLRR210 pKa = 11.84GYY212 pKa = 10.16GINIPDD218 pKa = 3.87KK219 pKa = 11.55DD220 pKa = 3.62EE221 pKa = 5.22DD222 pKa = 3.89KK223 pKa = 11.31PEE225 pKa = 5.63LIDD228 pKa = 4.85RR229 pKa = 11.84WKK231 pKa = 10.74DD232 pKa = 2.94WQYY235 pKa = 11.31PSNTINPATGAPSSACSWVFKK256 pKa = 10.77NGSRR260 pKa = 11.84DD261 pKa = 3.31PKK263 pKa = 10.41FFKK266 pKa = 10.86EE267 pKa = 3.76PGFLVGISVTRR278 pKa = 11.84PKK280 pKa = 11.03VYY282 pKa = 9.65MGNLSGSAASFAKK295 pKa = 10.29RR296 pKa = 11.84AWDD299 pKa = 3.23WMPNYY304 pKa = 10.48LMAYY308 pKa = 9.12PEE310 pKa = 4.21AALKK314 pKa = 10.47QFGVDD319 pKa = 3.51AGPLGVRR326 pKa = 11.84TTDD329 pKa = 3.04TDD331 pKa = 3.99SYY333 pKa = 9.32WIDD336 pKa = 3.33MRR338 pKa = 11.84DD339 pKa = 3.22EE340 pKa = 4.42LLYY343 pKa = 11.04GDD345 pKa = 4.51QFQNMHH351 pKa = 6.61AFDD354 pKa = 4.18AAGATANTDD363 pKa = 2.79GSNNLFGLPLTDD375 pKa = 4.18FTWKK379 pKa = 10.56YY380 pKa = 7.48PTEE383 pKa = 3.88AMINSLFVQDD393 pKa = 4.96DD394 pKa = 3.86GSGRR398 pKa = 11.84IRR400 pKa = 11.84QDD402 pKa = 2.94GYY404 pKa = 11.21VGLSIKK410 pKa = 9.97GAQGASPVDD419 pKa = 3.54YY420 pKa = 10.93TMGNIAEE427 pKa = 4.38TT428 pKa = 3.81
MM1 pKa = 7.29EE2 pKa = 5.9NIMRR6 pKa = 11.84TLMSAPPTGRR16 pKa = 11.84VGRR19 pKa = 11.84YY20 pKa = 6.92PKK22 pKa = 10.55HH23 pKa = 6.4PFLTQEE29 pKa = 4.55KK30 pKa = 9.31PFTAQPFFLARR41 pKa = 11.84VLPGEE46 pKa = 4.11TLQNLRR52 pKa = 11.84LEE54 pKa = 4.5SRR56 pKa = 11.84VITDD60 pKa = 4.82PIKK63 pKa = 10.91NSIIGWSKK71 pKa = 9.69EE72 pKa = 3.73YY73 pKa = 10.58YY74 pKa = 8.92VFYY77 pKa = 11.25VKK79 pKa = 10.76VSDD82 pKa = 5.08LMIDD86 pKa = 4.11AIKK89 pKa = 11.15DD90 pKa = 3.46MFIDD94 pKa = 3.67PTNAEE99 pKa = 3.89ITGQDD104 pKa = 2.85AATNVQRR111 pKa = 11.84TYY113 pKa = 10.3TPQGGIDD120 pKa = 3.54WTGMCLTRR128 pKa = 11.84IVEE131 pKa = 4.41TYY133 pKa = 10.38FRR135 pKa = 11.84DD136 pKa = 3.7EE137 pKa = 4.45GEE139 pKa = 4.33TAASHH144 pKa = 6.52ALADD148 pKa = 4.14GTPMVQIRR156 pKa = 11.84QNSFLDD162 pKa = 4.03SITDD166 pKa = 3.31KK167 pKa = 11.77DD168 pKa = 4.48EE169 pKa = 4.43IPEE172 pKa = 4.17GAAISTATDD181 pKa = 3.13AGDD184 pKa = 4.34LDD186 pKa = 4.13RR187 pKa = 11.84LMDD190 pKa = 4.35AFEE193 pKa = 4.39MARR196 pKa = 11.84SLGMANMSYY205 pKa = 11.15EE206 pKa = 4.15DD207 pKa = 3.71FLRR210 pKa = 11.84GYY212 pKa = 10.16GINIPDD218 pKa = 3.87KK219 pKa = 11.55DD220 pKa = 3.62EE221 pKa = 5.22DD222 pKa = 3.89KK223 pKa = 11.31PEE225 pKa = 5.63LIDD228 pKa = 4.85RR229 pKa = 11.84WKK231 pKa = 10.74DD232 pKa = 2.94WQYY235 pKa = 11.31PSNTINPATGAPSSACSWVFKK256 pKa = 10.77NGSRR260 pKa = 11.84DD261 pKa = 3.31PKK263 pKa = 10.41FFKK266 pKa = 10.86EE267 pKa = 3.76PGFLVGISVTRR278 pKa = 11.84PKK280 pKa = 11.03VYY282 pKa = 9.65MGNLSGSAASFAKK295 pKa = 10.29RR296 pKa = 11.84AWDD299 pKa = 3.23WMPNYY304 pKa = 10.48LMAYY308 pKa = 9.12PEE310 pKa = 4.21AALKK314 pKa = 10.47QFGVDD319 pKa = 3.51AGPLGVRR326 pKa = 11.84TTDD329 pKa = 3.04TDD331 pKa = 3.99SYY333 pKa = 9.32WIDD336 pKa = 3.33MRR338 pKa = 11.84DD339 pKa = 3.22EE340 pKa = 4.42LLYY343 pKa = 11.04GDD345 pKa = 4.51QFQNMHH351 pKa = 6.61AFDD354 pKa = 4.18AAGATANTDD363 pKa = 2.79GSNNLFGLPLTDD375 pKa = 4.18FTWKK379 pKa = 10.56YY380 pKa = 7.48PTEE383 pKa = 3.88AMINSLFVQDD393 pKa = 4.96DD394 pKa = 3.86GSGRR398 pKa = 11.84IRR400 pKa = 11.84QDD402 pKa = 2.94GYY404 pKa = 11.21VGLSIKK410 pKa = 9.97GAQGASPVDD419 pKa = 3.54YY420 pKa = 10.93TMGNIAEE427 pKa = 4.38TT428 pKa = 3.81
Molecular weight: 47.56 kDa
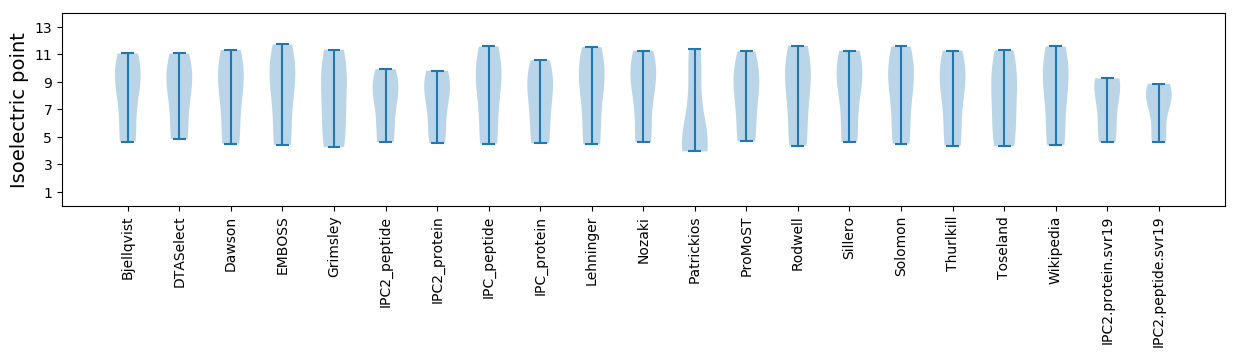
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4PJ67|A0A2H4PJ67_9VIRU Replication initiator OS=Citromicrobium phage vB_Cib_ssDNA_P1 OX=2053684 PE=4 SV=1
MM1 pKa = 7.39ARR3 pKa = 11.84DD4 pKa = 3.67DD5 pKa = 3.78ARR7 pKa = 11.84DD8 pKa = 3.74YY9 pKa = 11.44LWRR12 pKa = 11.84QQLGDD17 pKa = 3.5GHH19 pKa = 6.12NCSRR23 pKa = 11.84PVEE26 pKa = 4.18LPSIGLPSMLEE37 pKa = 3.87PDD39 pKa = 3.87RR40 pKa = 11.84TPLPVTIMARR50 pKa = 11.84CRR52 pKa = 11.84KK53 pKa = 9.81CPEE56 pKa = 4.04CLAHH60 pKa = 7.79RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84LWTARR68 pKa = 11.84AVDD71 pKa = 4.0EE72 pKa = 4.47VQASSRR78 pKa = 11.84TWFGTLTVAPAHH90 pKa = 6.3RR91 pKa = 11.84VRR93 pKa = 11.84LAYY96 pKa = 10.09RR97 pKa = 11.84AEE99 pKa = 4.09AKK101 pKa = 10.25VLRR104 pKa = 11.84AGGEE108 pKa = 4.34SLSSLDD114 pKa = 2.86RR115 pKa = 11.84SEE117 pKa = 4.81RR118 pKa = 11.84YY119 pKa = 9.5RR120 pKa = 11.84ILANEE125 pKa = 4.9LGQEE129 pKa = 3.96VTKK132 pKa = 9.77WLKK135 pKa = 10.17RR136 pKa = 11.84VRR138 pKa = 11.84KK139 pKa = 9.56QSGASLRR146 pKa = 11.84YY147 pKa = 9.44LLVAEE152 pKa = 4.37AHH154 pKa = 6.5KK155 pKa = 11.22SGDD158 pKa = 3.35PHH160 pKa = 5.22MHH162 pKa = 6.38ILVHH166 pKa = 6.68EE167 pKa = 5.36DD168 pKa = 3.16GDD170 pKa = 4.28PVTKK174 pKa = 10.54RR175 pKa = 11.84MLQDD179 pKa = 2.61QWRR182 pKa = 11.84IGFSQWKK189 pKa = 9.79LVDD192 pKa = 3.75QDD194 pKa = 3.51PGAAVYY200 pKa = 9.38VCKK203 pKa = 10.78YY204 pKa = 9.0LAKK207 pKa = 10.46DD208 pKa = 2.76ALTRR212 pKa = 11.84VRR214 pKa = 11.84ASRR217 pKa = 11.84RR218 pKa = 11.84YY219 pKa = 8.68GQPQLVRR226 pKa = 11.84SLTEE230 pKa = 4.6RR231 pKa = 11.84IHH233 pKa = 6.92KK234 pKa = 9.62MRR236 pKa = 11.84DD237 pKa = 3.2AVHH240 pKa = 6.0EE241 pKa = 4.28ASSRR245 pKa = 11.84RR246 pKa = 11.84EE247 pKa = 3.54PDD249 pKa = 2.73
MM1 pKa = 7.39ARR3 pKa = 11.84DD4 pKa = 3.67DD5 pKa = 3.78ARR7 pKa = 11.84DD8 pKa = 3.74YY9 pKa = 11.44LWRR12 pKa = 11.84QQLGDD17 pKa = 3.5GHH19 pKa = 6.12NCSRR23 pKa = 11.84PVEE26 pKa = 4.18LPSIGLPSMLEE37 pKa = 3.87PDD39 pKa = 3.87RR40 pKa = 11.84TPLPVTIMARR50 pKa = 11.84CRR52 pKa = 11.84KK53 pKa = 9.81CPEE56 pKa = 4.04CLAHH60 pKa = 7.79RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84LWTARR68 pKa = 11.84AVDD71 pKa = 4.0EE72 pKa = 4.47VQASSRR78 pKa = 11.84TWFGTLTVAPAHH90 pKa = 6.3RR91 pKa = 11.84VRR93 pKa = 11.84LAYY96 pKa = 10.09RR97 pKa = 11.84AEE99 pKa = 4.09AKK101 pKa = 10.25VLRR104 pKa = 11.84AGGEE108 pKa = 4.34SLSSLDD114 pKa = 2.86RR115 pKa = 11.84SEE117 pKa = 4.81RR118 pKa = 11.84YY119 pKa = 9.5RR120 pKa = 11.84ILANEE125 pKa = 4.9LGQEE129 pKa = 3.96VTKK132 pKa = 9.77WLKK135 pKa = 10.17RR136 pKa = 11.84VRR138 pKa = 11.84KK139 pKa = 9.56QSGASLRR146 pKa = 11.84YY147 pKa = 9.44LLVAEE152 pKa = 4.37AHH154 pKa = 6.5KK155 pKa = 11.22SGDD158 pKa = 3.35PHH160 pKa = 5.22MHH162 pKa = 6.38ILVHH166 pKa = 6.68EE167 pKa = 5.36DD168 pKa = 3.16GDD170 pKa = 4.28PVTKK174 pKa = 10.54RR175 pKa = 11.84MLQDD179 pKa = 2.61QWRR182 pKa = 11.84IGFSQWKK189 pKa = 9.79LVDD192 pKa = 3.75QDD194 pKa = 3.51PGAAVYY200 pKa = 9.38VCKK203 pKa = 10.78YY204 pKa = 9.0LAKK207 pKa = 10.46DD208 pKa = 2.76ALTRR212 pKa = 11.84VRR214 pKa = 11.84ASRR217 pKa = 11.84RR218 pKa = 11.84YY219 pKa = 8.68GQPQLVRR226 pKa = 11.84SLTEE230 pKa = 4.6RR231 pKa = 11.84IHH233 pKa = 6.92KK234 pKa = 9.62MRR236 pKa = 11.84DD237 pKa = 3.2AVHH240 pKa = 6.0EE241 pKa = 4.28ASSRR245 pKa = 11.84RR246 pKa = 11.84EE247 pKa = 3.54PDD249 pKa = 2.73
Molecular weight: 28.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1322 |
37 |
428 |
220.3 |
24.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.682 ± 1.248 | 1.286 ± 0.388 |
5.9 ± 1.157 | 5.446 ± 0.691 |
3.48 ± 0.95 | 7.867 ± 1.11 |
1.815 ± 0.584 | 5.144 ± 0.726 |
4.614 ± 0.719 | 7.564 ± 0.792 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.177 ± 0.451 | 3.48 ± 0.789 |
8.169 ± 1.548 | 3.177 ± 0.457 |
7.262 ± 1.566 | 5.673 ± 0.651 |
5.825 ± 0.697 | 5.144 ± 1.43 |
2.118 ± 0.32 | 3.177 ± 0.533 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
