
Fibrella sp. ES10-3-2-2
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Fibrella; unclassified Fibrella
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
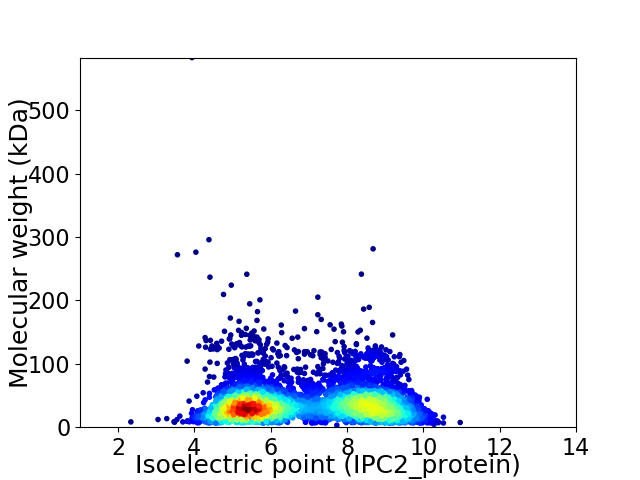
Virtual 2D-PAGE plot for 5038 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6E758|A0A1W6E758_9BACT Peptidase C69 OS=Fibrella sp. ES10-3-2-2 OX=1834519 GN=A6C57_12990 PE=3 SV=1
MM1 pKa = 7.67LYY3 pKa = 10.16YY4 pKa = 10.72VKK6 pKa = 9.94TYY8 pKa = 9.96FLVGMRR14 pKa = 11.84PIVLVWALLFGLLSTAQTNAQTLPCNYY41 pKa = 7.8RR42 pKa = 11.84TLTTNNYY49 pKa = 9.63SGAFIVNSRR58 pKa = 11.84TGGTGGFLGIGAGNVTNEE76 pKa = 3.85GRR78 pKa = 11.84VIDD81 pKa = 4.39ADD83 pKa = 4.11LNNFASINTTLTLASGGEE101 pKa = 3.92ISVDD105 pKa = 3.02AGGTVVFGAGATAGYY120 pKa = 10.16VVDD123 pKa = 4.92GSSGLGANLLGSVTIITYY141 pKa = 10.81LNGIEE146 pKa = 4.03QEE148 pKa = 4.13RR149 pKa = 11.84SNSSSLLDD157 pKa = 3.59LVLLSGSGRR166 pKa = 11.84RR167 pKa = 11.84TLGFVTTKK175 pKa = 10.75NFDD178 pKa = 3.35EE179 pKa = 4.33VQIRR183 pKa = 11.84VSSLISLLTSLPVYY197 pKa = 10.68YY198 pKa = 10.13PFVQYY203 pKa = 8.75KK204 pKa = 8.76TLGATATATSASGLTTADD222 pKa = 3.07GSVALAVTGGRR233 pKa = 11.84APFTYY238 pKa = 9.89LWSNGATTANLTNVLPGTYY257 pKa = 9.91SVTVTDD263 pKa = 5.34ANGCTTTASATVGIKK278 pKa = 9.85VAACPVPGQNGFTKK292 pKa = 10.57FSFATAPTITGQGVGRR308 pKa = 11.84KK309 pKa = 8.77GRR311 pKa = 11.84YY312 pKa = 8.81VNVASIGGVAVDD324 pKa = 4.48VIGEE328 pKa = 4.26VITYY332 pKa = 10.37SGTADD337 pKa = 3.02ATFPRR342 pKa = 11.84FDD344 pKa = 4.04NFSSTSGAFLARR356 pKa = 11.84YY357 pKa = 8.62AISGASTAPAGLTSTVRR374 pKa = 11.84WSVVKK379 pKa = 9.9TGTNLPVPFQGAFTVGDD396 pKa = 3.43IDD398 pKa = 5.37NDD400 pKa = 3.48VTLNSTTLEE409 pKa = 4.1SIIVNKK415 pKa = 10.31SDD417 pKa = 3.31LYY419 pKa = 10.82SYY421 pKa = 10.88KK422 pKa = 10.52LSSPTNTSVISSSASPTIKK441 pKa = 10.58FQGTLNQVGIDD452 pKa = 3.79GVDD455 pKa = 3.31PAFTVALAYY464 pKa = 10.62VGVSSFEE471 pKa = 3.6VTYY474 pKa = 10.94AKK476 pKa = 10.49VGSSSGTANFPFDD489 pKa = 3.81GEE491 pKa = 4.3GGIVFLASTCTPVLDD506 pKa = 3.87TDD508 pKa = 4.06GDD510 pKa = 4.31GVANAIDD517 pKa = 3.91TDD519 pKa = 4.09DD520 pKa = 5.29DD521 pKa = 4.32NDD523 pKa = 5.04GILDD527 pKa = 3.8DD528 pKa = 4.16TEE530 pKa = 5.75GGVFVDD536 pKa = 4.19SDD538 pKa = 3.85GDD540 pKa = 4.27GISNALDD547 pKa = 3.95LDD549 pKa = 3.94SDD551 pKa = 4.27GDD553 pKa = 4.26GIPDD557 pKa = 4.04NIEE560 pKa = 3.83AQTTAGYY567 pKa = 9.26IAPGTAVDD575 pKa = 3.65AAGLLTSYY583 pKa = 9.25TATNGLIPVNTDD595 pKa = 2.57GTDD598 pKa = 3.09TADD601 pKa = 4.29YY602 pKa = 11.23LDD604 pKa = 5.3LDD606 pKa = 4.08SDD608 pKa = 3.79NDD610 pKa = 3.66TRR612 pKa = 11.84TDD614 pKa = 3.35TVEE617 pKa = 4.69AGITLANADD626 pKa = 4.39DD627 pKa = 5.38DD628 pKa = 5.92KK629 pKa = 11.84DD630 pKa = 4.08GLDD633 pKa = 4.16NSPDD637 pKa = 3.59TNDD640 pKa = 3.51NLFGPVNAGITNPLTFYY657 pKa = 10.15PNNGTEE663 pKa = 4.14VLWRR667 pKa = 11.84VKK669 pKa = 10.37RR670 pKa = 11.84GAFTYY675 pKa = 9.93GNCALATFSGQFVVGIPSSGVLTIPITTSIDD706 pKa = 3.19GAITITSVSGSGISSVPASVTAILIAGQTTLSIPIAYY743 pKa = 9.48DD744 pKa = 3.24GSGVIGTRR752 pKa = 11.84NLTVSSVDD760 pKa = 3.23ATGTCAPIVAVIGLADD776 pKa = 4.06LTTSIGIPVPTLIGAQTSSLPISVSNIGSAATTGPITTTLTLPASVTAPATFTSNGFGCTTTGTSVSCTSSAILANGSSTTFAVPITPPLATVGTTLSFTNTVMTTQEE884 pKa = 3.38ISVTNNTGTSTALVTGAPDD903 pKa = 3.56LAVSIGQPSPALVVAQTSTIPVSVSNVGTIPTIGPITVTLTIPASVTAPATFTNNGFGCSTSGGTITCTSAGPIANAASLTFGIPVTPLSAALGTTPTFSGLVATTGDD1011 pKa = 3.8IITANNSATMTANTAVACAVGSAIPILKK1039 pKa = 10.25
MM1 pKa = 7.67LYY3 pKa = 10.16YY4 pKa = 10.72VKK6 pKa = 9.94TYY8 pKa = 9.96FLVGMRR14 pKa = 11.84PIVLVWALLFGLLSTAQTNAQTLPCNYY41 pKa = 7.8RR42 pKa = 11.84TLTTNNYY49 pKa = 9.63SGAFIVNSRR58 pKa = 11.84TGGTGGFLGIGAGNVTNEE76 pKa = 3.85GRR78 pKa = 11.84VIDD81 pKa = 4.39ADD83 pKa = 4.11LNNFASINTTLTLASGGEE101 pKa = 3.92ISVDD105 pKa = 3.02AGGTVVFGAGATAGYY120 pKa = 10.16VVDD123 pKa = 4.92GSSGLGANLLGSVTIITYY141 pKa = 10.81LNGIEE146 pKa = 4.03QEE148 pKa = 4.13RR149 pKa = 11.84SNSSSLLDD157 pKa = 3.59LVLLSGSGRR166 pKa = 11.84RR167 pKa = 11.84TLGFVTTKK175 pKa = 10.75NFDD178 pKa = 3.35EE179 pKa = 4.33VQIRR183 pKa = 11.84VSSLISLLTSLPVYY197 pKa = 10.68YY198 pKa = 10.13PFVQYY203 pKa = 8.75KK204 pKa = 8.76TLGATATATSASGLTTADD222 pKa = 3.07GSVALAVTGGRR233 pKa = 11.84APFTYY238 pKa = 9.89LWSNGATTANLTNVLPGTYY257 pKa = 9.91SVTVTDD263 pKa = 5.34ANGCTTTASATVGIKK278 pKa = 9.85VAACPVPGQNGFTKK292 pKa = 10.57FSFATAPTITGQGVGRR308 pKa = 11.84KK309 pKa = 8.77GRR311 pKa = 11.84YY312 pKa = 8.81VNVASIGGVAVDD324 pKa = 4.48VIGEE328 pKa = 4.26VITYY332 pKa = 10.37SGTADD337 pKa = 3.02ATFPRR342 pKa = 11.84FDD344 pKa = 4.04NFSSTSGAFLARR356 pKa = 11.84YY357 pKa = 8.62AISGASTAPAGLTSTVRR374 pKa = 11.84WSVVKK379 pKa = 9.9TGTNLPVPFQGAFTVGDD396 pKa = 3.43IDD398 pKa = 5.37NDD400 pKa = 3.48VTLNSTTLEE409 pKa = 4.1SIIVNKK415 pKa = 10.31SDD417 pKa = 3.31LYY419 pKa = 10.82SYY421 pKa = 10.88KK422 pKa = 10.52LSSPTNTSVISSSASPTIKK441 pKa = 10.58FQGTLNQVGIDD452 pKa = 3.79GVDD455 pKa = 3.31PAFTVALAYY464 pKa = 10.62VGVSSFEE471 pKa = 3.6VTYY474 pKa = 10.94AKK476 pKa = 10.49VGSSSGTANFPFDD489 pKa = 3.81GEE491 pKa = 4.3GGIVFLASTCTPVLDD506 pKa = 3.87TDD508 pKa = 4.06GDD510 pKa = 4.31GVANAIDD517 pKa = 3.91TDD519 pKa = 4.09DD520 pKa = 5.29DD521 pKa = 4.32NDD523 pKa = 5.04GILDD527 pKa = 3.8DD528 pKa = 4.16TEE530 pKa = 5.75GGVFVDD536 pKa = 4.19SDD538 pKa = 3.85GDD540 pKa = 4.27GISNALDD547 pKa = 3.95LDD549 pKa = 3.94SDD551 pKa = 4.27GDD553 pKa = 4.26GIPDD557 pKa = 4.04NIEE560 pKa = 3.83AQTTAGYY567 pKa = 9.26IAPGTAVDD575 pKa = 3.65AAGLLTSYY583 pKa = 9.25TATNGLIPVNTDD595 pKa = 2.57GTDD598 pKa = 3.09TADD601 pKa = 4.29YY602 pKa = 11.23LDD604 pKa = 5.3LDD606 pKa = 4.08SDD608 pKa = 3.79NDD610 pKa = 3.66TRR612 pKa = 11.84TDD614 pKa = 3.35TVEE617 pKa = 4.69AGITLANADD626 pKa = 4.39DD627 pKa = 5.38DD628 pKa = 5.92KK629 pKa = 11.84DD630 pKa = 4.08GLDD633 pKa = 4.16NSPDD637 pKa = 3.59TNDD640 pKa = 3.51NLFGPVNAGITNPLTFYY657 pKa = 10.15PNNGTEE663 pKa = 4.14VLWRR667 pKa = 11.84VKK669 pKa = 10.37RR670 pKa = 11.84GAFTYY675 pKa = 9.93GNCALATFSGQFVVGIPSSGVLTIPITTSIDD706 pKa = 3.19GAITITSVSGSGISSVPASVTAILIAGQTTLSIPIAYY743 pKa = 9.48DD744 pKa = 3.24GSGVIGTRR752 pKa = 11.84NLTVSSVDD760 pKa = 3.23ATGTCAPIVAVIGLADD776 pKa = 4.06LTTSIGIPVPTLIGAQTSSLPISVSNIGSAATTGPITTTLTLPASVTAPATFTSNGFGCTTTGTSVSCTSSAILANGSSTTFAVPITPPLATVGTTLSFTNTVMTTQEE884 pKa = 3.38ISVTNNTGTSTALVTGAPDD903 pKa = 3.56LAVSIGQPSPALVVAQTSTIPVSVSNVGTIPTIGPITVTLTIPASVTAPATFTNNGFGCSTSGGTITCTSAGPIANAASLTFGIPVTPLSAALGTTPTFSGLVATTGDD1011 pKa = 3.8IITANNSATMTANTAVACAVGSAIPILKK1039 pKa = 10.25
Molecular weight: 104.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6E9W9|A0A1W6E9W9_9BACT DUF4918 domain-containing protein OS=Fibrella sp. ES10-3-2-2 OX=1834519 GN=A6C57_18830 PE=4 SV=1
MM1 pKa = 7.98PKK3 pKa = 10.22QKK5 pKa = 9.71TNSAAKK11 pKa = 10.06KK12 pKa = 9.44RR13 pKa = 11.84FKK15 pKa = 10.53LTGTGKK21 pKa = 10.6LKK23 pKa = 10.41RR24 pKa = 11.84KK25 pKa = 8.99HH26 pKa = 6.27AFHH29 pKa = 6.83SHH31 pKa = 5.99ILTKK35 pKa = 9.89KK36 pKa = 4.76TTKK39 pKa = 9.74QKK41 pKa = 10.98RR42 pKa = 11.84NLAHH46 pKa = 6.91SSTVHH51 pKa = 5.26PTNMTRR57 pKa = 11.84IKK59 pKa = 10.88ALLRR63 pKa = 11.84LSS65 pKa = 3.59
MM1 pKa = 7.98PKK3 pKa = 10.22QKK5 pKa = 9.71TNSAAKK11 pKa = 10.06KK12 pKa = 9.44RR13 pKa = 11.84FKK15 pKa = 10.53LTGTGKK21 pKa = 10.6LKK23 pKa = 10.41RR24 pKa = 11.84KK25 pKa = 8.99HH26 pKa = 6.27AFHH29 pKa = 6.83SHH31 pKa = 5.99ILTKK35 pKa = 9.89KK36 pKa = 4.76TTKK39 pKa = 9.74QKK41 pKa = 10.98RR42 pKa = 11.84NLAHH46 pKa = 6.91SSTVHH51 pKa = 5.26PTNMTRR57 pKa = 11.84IKK59 pKa = 10.88ALLRR63 pKa = 11.84LSS65 pKa = 3.59
Molecular weight: 7.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1868441 |
26 |
5706 |
370.9 |
41.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.967 ± 0.04 | 0.742 ± 0.013 |
5.343 ± 0.027 | 4.816 ± 0.03 |
4.282 ± 0.023 | 7.482 ± 0.031 |
1.744 ± 0.017 | 5.444 ± 0.027 |
4.56 ± 0.034 | 10.111 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.132 ± 0.016 | 4.523 ± 0.033 |
4.744 ± 0.023 | 4.395 ± 0.025 |
5.415 ± 0.032 | 6.123 ± 0.034 |
7.006 ± 0.054 | 7.105 ± 0.027 |
1.314 ± 0.015 | 3.753 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
