
Candidatus Vecturithrix granuli
Taxonomy: cellular organisms; Bacteria; Bacteria incertae sedis; Candidatus Vecturithrix
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
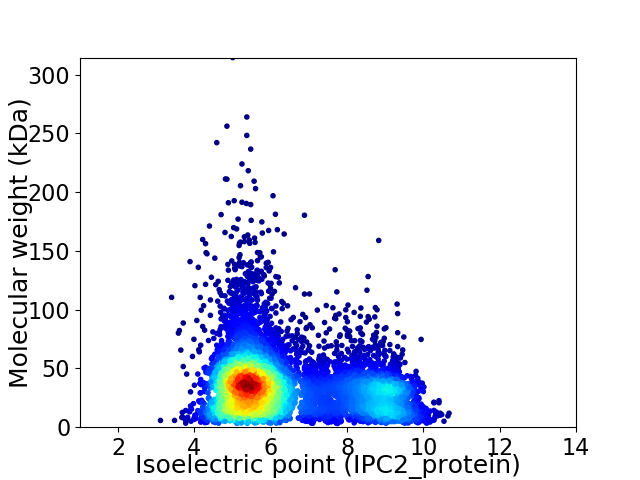
Virtual 2D-PAGE plot for 7048 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A081BUC5|A0A081BUC5_9BACT Multicomponent Na+:H+ antiporter subunit B OS=Candidatus Vecturithrix granuli OX=1499967 GN=U27_02891 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 10.33RR3 pKa = 11.84QVNRR7 pKa = 11.84QLLYY11 pKa = 9.05MHH13 pKa = 7.34IFSKK17 pKa = 10.02TISFVLLLGIVGAVLIGIGGCKK39 pKa = 9.27GVNVNPTTPTIDD51 pKa = 3.39TPTPEE56 pKa = 4.78PEE58 pKa = 4.14STATPDD64 pKa = 3.43GGEE67 pKa = 4.12STPTPEE73 pKa = 6.16GEE75 pKa = 4.16GCDD78 pKa = 3.94NYY80 pKa = 11.44GEE82 pKa = 4.61TPDD85 pKa = 4.46DD86 pKa = 4.24PNLIVLMPDD95 pKa = 3.56GDD97 pKa = 4.04PVAISMDD104 pKa = 3.86CTDD107 pKa = 5.31DD108 pKa = 3.58IDD110 pKa = 3.59WYY112 pKa = 10.69KK113 pKa = 10.6IEE115 pKa = 4.18ITQSPVTLEE124 pKa = 3.77IKK126 pKa = 9.37LTNLPGNSDD135 pKa = 3.99YY136 pKa = 11.82DD137 pKa = 4.44LIAYY141 pKa = 7.93DD142 pKa = 3.96ANLLEE147 pKa = 4.37MEE149 pKa = 4.87NGRR152 pKa = 11.84SAQTGNVDD160 pKa = 2.81EE161 pKa = 4.58HH162 pKa = 6.98LRR164 pKa = 11.84LNVTDD169 pKa = 4.59FMIYY173 pKa = 10.31LQIHH177 pKa = 6.43SYY179 pKa = 9.65NGKK182 pKa = 7.94GTAILRR188 pKa = 11.84ISIVDD193 pKa = 3.83DD194 pKa = 4.17SPVEE198 pKa = 4.27PGDD201 pKa = 5.55DD202 pKa = 5.02DD203 pKa = 6.88DD204 pKa = 7.59GDD206 pKa = 5.58DD207 pKa = 4.62DD208 pKa = 6.54DD209 pKa = 6.93GDD211 pKa = 3.83EE212 pKa = 4.75FEE214 pKa = 4.27QLSYY218 pKa = 11.57EE219 pKa = 4.64DD220 pKa = 4.79ILFQEE225 pKa = 4.87LSSYY229 pKa = 10.6PRR231 pKa = 11.84GTMSSLLEE239 pKa = 3.94RR240 pKa = 11.84SVEE243 pKa = 4.12TTITKK248 pKa = 10.2SVFCQLTDD256 pKa = 3.39SQLTGTFTIGNYY268 pKa = 10.06AHH270 pKa = 6.55VEE272 pKa = 3.84RR273 pKa = 11.84DD274 pKa = 3.43FLQSVDD280 pKa = 5.06YY281 pKa = 10.75IDD283 pKa = 4.04GWSLVVFNGSKK294 pKa = 10.51EE295 pKa = 4.03LLDD298 pKa = 4.07NLTLTIRR305 pKa = 11.84VTLYY309 pKa = 10.83APDD312 pKa = 3.38VGLEE316 pKa = 3.96VEE318 pKa = 4.54VPYY321 pKa = 11.44NMEE324 pKa = 3.95IDD326 pKa = 3.82GNDD329 pKa = 3.51YY330 pKa = 9.46LTSTSEE336 pKa = 3.58NDD338 pKa = 3.22VYY340 pKa = 11.53YY341 pKa = 10.99LSQKK345 pKa = 10.83YY346 pKa = 10.35GDD348 pKa = 3.82LAGNSSSDD356 pKa = 3.03IRR358 pKa = 11.84ITSGVVGDD366 pKa = 4.75IGDD369 pKa = 4.38LFANNIFTQQVEE381 pKa = 4.61VSWEE385 pKa = 3.97YY386 pKa = 9.95TDD388 pKa = 4.86EE389 pKa = 4.57SGGYY393 pKa = 9.76CSGRR397 pKa = 11.84VSGRR401 pKa = 11.84SIVDD405 pKa = 3.84FAALSIRR412 pKa = 3.82
MM1 pKa = 7.33KK2 pKa = 10.33RR3 pKa = 11.84QVNRR7 pKa = 11.84QLLYY11 pKa = 9.05MHH13 pKa = 7.34IFSKK17 pKa = 10.02TISFVLLLGIVGAVLIGIGGCKK39 pKa = 9.27GVNVNPTTPTIDD51 pKa = 3.39TPTPEE56 pKa = 4.78PEE58 pKa = 4.14STATPDD64 pKa = 3.43GGEE67 pKa = 4.12STPTPEE73 pKa = 6.16GEE75 pKa = 4.16GCDD78 pKa = 3.94NYY80 pKa = 11.44GEE82 pKa = 4.61TPDD85 pKa = 4.46DD86 pKa = 4.24PNLIVLMPDD95 pKa = 3.56GDD97 pKa = 4.04PVAISMDD104 pKa = 3.86CTDD107 pKa = 5.31DD108 pKa = 3.58IDD110 pKa = 3.59WYY112 pKa = 10.69KK113 pKa = 10.6IEE115 pKa = 4.18ITQSPVTLEE124 pKa = 3.77IKK126 pKa = 9.37LTNLPGNSDD135 pKa = 3.99YY136 pKa = 11.82DD137 pKa = 4.44LIAYY141 pKa = 7.93DD142 pKa = 3.96ANLLEE147 pKa = 4.37MEE149 pKa = 4.87NGRR152 pKa = 11.84SAQTGNVDD160 pKa = 2.81EE161 pKa = 4.58HH162 pKa = 6.98LRR164 pKa = 11.84LNVTDD169 pKa = 4.59FMIYY173 pKa = 10.31LQIHH177 pKa = 6.43SYY179 pKa = 9.65NGKK182 pKa = 7.94GTAILRR188 pKa = 11.84ISIVDD193 pKa = 3.83DD194 pKa = 4.17SPVEE198 pKa = 4.27PGDD201 pKa = 5.55DD202 pKa = 5.02DD203 pKa = 6.88DD204 pKa = 7.59GDD206 pKa = 5.58DD207 pKa = 4.62DD208 pKa = 6.54DD209 pKa = 6.93GDD211 pKa = 3.83EE212 pKa = 4.75FEE214 pKa = 4.27QLSYY218 pKa = 11.57EE219 pKa = 4.64DD220 pKa = 4.79ILFQEE225 pKa = 4.87LSSYY229 pKa = 10.6PRR231 pKa = 11.84GTMSSLLEE239 pKa = 3.94RR240 pKa = 11.84SVEE243 pKa = 4.12TTITKK248 pKa = 10.2SVFCQLTDD256 pKa = 3.39SQLTGTFTIGNYY268 pKa = 10.06AHH270 pKa = 6.55VEE272 pKa = 3.84RR273 pKa = 11.84DD274 pKa = 3.43FLQSVDD280 pKa = 5.06YY281 pKa = 10.75IDD283 pKa = 4.04GWSLVVFNGSKK294 pKa = 10.51EE295 pKa = 4.03LLDD298 pKa = 4.07NLTLTIRR305 pKa = 11.84VTLYY309 pKa = 10.83APDD312 pKa = 3.38VGLEE316 pKa = 3.96VEE318 pKa = 4.54VPYY321 pKa = 11.44NMEE324 pKa = 3.95IDD326 pKa = 3.82GNDD329 pKa = 3.51YY330 pKa = 9.46LTSTSEE336 pKa = 3.58NDD338 pKa = 3.22VYY340 pKa = 11.53YY341 pKa = 10.99LSQKK345 pKa = 10.83YY346 pKa = 10.35GDD348 pKa = 3.82LAGNSSSDD356 pKa = 3.03IRR358 pKa = 11.84ITSGVVGDD366 pKa = 4.75IGDD369 pKa = 4.38LFANNIFTQQVEE381 pKa = 4.61VSWEE385 pKa = 3.97YY386 pKa = 9.95TDD388 pKa = 4.86EE389 pKa = 4.57SGGYY393 pKa = 9.76CSGRR397 pKa = 11.84VSGRR401 pKa = 11.84SIVDD405 pKa = 3.84FAALSIRR412 pKa = 3.82
Molecular weight: 45.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A081BXK5|A0A081BXK5_9BACT HEPN_Swt1 domain-containing protein OS=Candidatus Vecturithrix granuli OX=1499967 GN=U27_04024 PE=4 SV=1
MM1 pKa = 6.9EE2 pKa = 4.6TVDD5 pKa = 3.85EE6 pKa = 4.4RR7 pKa = 11.84TGSSRR12 pKa = 11.84SRR14 pKa = 11.84QTVWPSLRR22 pKa = 11.84IPQASRR28 pKa = 11.84DD29 pKa = 3.8WASTTAAYY37 pKa = 9.78RR38 pKa = 11.84LCANPKK44 pKa = 9.27VSAARR49 pKa = 11.84ILEE52 pKa = 3.84PHH54 pKa = 5.76QRR56 pKa = 11.84QTCRR60 pKa = 11.84RR61 pKa = 11.84MAGEE65 pKa = 4.05SRR67 pKa = 11.84ILAGQEE73 pKa = 4.04TCFLNYY79 pKa = 9.16TSHH82 pKa = 7.11PTTEE86 pKa = 4.05TGQMRR91 pKa = 11.84LVLRR95 pKa = 11.84ARR97 pKa = 11.84PVSGYY102 pKa = 7.71VTVAIPAQPQRR113 pKa = 11.84PARR116 pKa = 11.84TATVEE121 pKa = 3.77VRR123 pKa = 11.84YY124 pKa = 10.75GEE126 pKa = 4.29VTLRR130 pKa = 11.84PPSRR134 pKa = 11.84DD135 pKa = 3.29PSCQADD141 pKa = 4.13LRR143 pKa = 11.84PLTLAVVWARR153 pKa = 11.84EE154 pKa = 4.11VEE156 pKa = 4.07ASGDD160 pKa = 3.59VATPLEE166 pKa = 4.5WLLMTTVSIQTFLDD180 pKa = 3.2AVEE183 pKa = 3.92RR184 pKa = 11.84MRR186 pKa = 11.84WYY188 pKa = 10.3RR189 pKa = 11.84LRR191 pKa = 11.84WHH193 pKa = 6.88IEE195 pKa = 3.51IFHH198 pKa = 6.63KK199 pKa = 10.51VLKK202 pKa = 9.64SGCRR206 pKa = 11.84IADD209 pKa = 3.41YY210 pKa = 11.05RR211 pKa = 11.84LATVDD216 pKa = 3.37KK217 pKa = 10.19LVRR220 pKa = 11.84SVTLKK225 pKa = 10.97SVVAWQMQWMGPLHH239 pKa = 5.96RR240 pKa = 11.84TSTLPEE246 pKa = 4.09AVPTVHH252 pKa = 6.84QMVRR256 pKa = 11.84WIAKK260 pKa = 9.72LGGFLGRR267 pKa = 11.84KK268 pKa = 8.77HH269 pKa = 6.62DD270 pKa = 4.4RR271 pKa = 11.84EE272 pKa = 4.17PGGAGNGFTISPPCGTCCMSPHH294 pKa = 4.94LHH296 pKa = 6.35KK297 pKa = 10.87RR298 pKa = 11.84GGYY301 pKa = 8.55PGGRR305 pKa = 11.84NTGRR309 pKa = 11.84SRR311 pKa = 11.84GRR313 pKa = 11.84VCFSVEE319 pKa = 3.6QYY321 pKa = 10.13VRR323 pKa = 11.84IMIIRR328 pKa = 3.91
MM1 pKa = 6.9EE2 pKa = 4.6TVDD5 pKa = 3.85EE6 pKa = 4.4RR7 pKa = 11.84TGSSRR12 pKa = 11.84SRR14 pKa = 11.84QTVWPSLRR22 pKa = 11.84IPQASRR28 pKa = 11.84DD29 pKa = 3.8WASTTAAYY37 pKa = 9.78RR38 pKa = 11.84LCANPKK44 pKa = 9.27VSAARR49 pKa = 11.84ILEE52 pKa = 3.84PHH54 pKa = 5.76QRR56 pKa = 11.84QTCRR60 pKa = 11.84RR61 pKa = 11.84MAGEE65 pKa = 4.05SRR67 pKa = 11.84ILAGQEE73 pKa = 4.04TCFLNYY79 pKa = 9.16TSHH82 pKa = 7.11PTTEE86 pKa = 4.05TGQMRR91 pKa = 11.84LVLRR95 pKa = 11.84ARR97 pKa = 11.84PVSGYY102 pKa = 7.71VTVAIPAQPQRR113 pKa = 11.84PARR116 pKa = 11.84TATVEE121 pKa = 3.77VRR123 pKa = 11.84YY124 pKa = 10.75GEE126 pKa = 4.29VTLRR130 pKa = 11.84PPSRR134 pKa = 11.84DD135 pKa = 3.29PSCQADD141 pKa = 4.13LRR143 pKa = 11.84PLTLAVVWARR153 pKa = 11.84EE154 pKa = 4.11VEE156 pKa = 4.07ASGDD160 pKa = 3.59VATPLEE166 pKa = 4.5WLLMTTVSIQTFLDD180 pKa = 3.2AVEE183 pKa = 3.92RR184 pKa = 11.84MRR186 pKa = 11.84WYY188 pKa = 10.3RR189 pKa = 11.84LRR191 pKa = 11.84WHH193 pKa = 6.88IEE195 pKa = 3.51IFHH198 pKa = 6.63KK199 pKa = 10.51VLKK202 pKa = 9.64SGCRR206 pKa = 11.84IADD209 pKa = 3.41YY210 pKa = 11.05RR211 pKa = 11.84LATVDD216 pKa = 3.37KK217 pKa = 10.19LVRR220 pKa = 11.84SVTLKK225 pKa = 10.97SVVAWQMQWMGPLHH239 pKa = 5.96RR240 pKa = 11.84TSTLPEE246 pKa = 4.09AVPTVHH252 pKa = 6.84QMVRR256 pKa = 11.84WIAKK260 pKa = 9.72LGGFLGRR267 pKa = 11.84KK268 pKa = 8.77HH269 pKa = 6.62DD270 pKa = 4.4RR271 pKa = 11.84EE272 pKa = 4.17PGGAGNGFTISPPCGTCCMSPHH294 pKa = 4.94LHH296 pKa = 6.35KK297 pKa = 10.87RR298 pKa = 11.84GGYY301 pKa = 8.55PGGRR305 pKa = 11.84NTGRR309 pKa = 11.84SRR311 pKa = 11.84GRR313 pKa = 11.84VCFSVEE319 pKa = 3.6QYY321 pKa = 10.13VRR323 pKa = 11.84IMIIRR328 pKa = 3.91
Molecular weight: 36.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2347583 |
29 |
2850 |
333.1 |
37.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.031 ± 0.027 | 1.103 ± 0.011 |
4.922 ± 0.022 | 6.774 ± 0.034 |
4.553 ± 0.023 | 6.635 ± 0.028 |
2.357 ± 0.014 | 7.188 ± 0.028 |
5.041 ± 0.029 | 10.513 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.494 ± 0.014 | 3.523 ± 0.016 |
4.353 ± 0.019 | 4.792 ± 0.026 |
5.186 ± 0.021 | 5.546 ± 0.021 |
5.574 ± 0.022 | 6.713 ± 0.024 |
1.235 ± 0.012 | 3.469 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
