
Clostridium saccharolyticum (strain ATCC 35040 / DSM 2544 / NRCC 2533 / WM1)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Lacrimispora; Lacrimispora saccharolytica
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
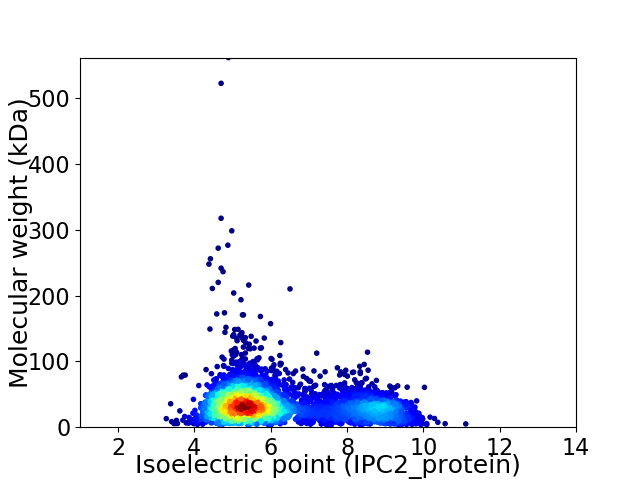
Virtual 2D-PAGE plot for 4133 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D9RA59|D9RA59_CLOSW Uncharacterized protein OS=Clostridium saccharolyticum (strain ATCC 35040 / DSM 2544 / NRCC 2533 / WM1) OX=610130 GN=Closa_3506 PE=4 SV=1
MM1 pKa = 7.46NKK3 pKa = 9.88RR4 pKa = 11.84LFSMFLALCMIVTMLPVSAMAEE26 pKa = 4.26EE27 pKa = 4.05IHH29 pKa = 5.41TTIGGSGEE37 pKa = 4.23IISFAPLTEE46 pKa = 3.91TAKK49 pKa = 10.73AVSLGTSIEE58 pKa = 4.06DD59 pKa = 3.61LEE61 pKa = 4.78LPEE64 pKa = 4.5TLTATVRR71 pKa = 11.84TAVPADD77 pKa = 3.72EE78 pKa = 6.49DD79 pKa = 3.99STQDD83 pKa = 3.12SGSPEE88 pKa = 4.04TATPTTAAEE97 pKa = 4.45PKK99 pKa = 9.12WKK101 pKa = 8.4EE102 pKa = 3.99TTGDD106 pKa = 3.89IPVEE110 pKa = 4.1WASPDD115 pKa = 3.27YY116 pKa = 11.79DD117 pKa = 3.45MDD119 pKa = 4.07TEE121 pKa = 4.41GVYY124 pKa = 10.7VFTPVIEE131 pKa = 5.12GYY133 pKa = 8.18TVSAPLPEE141 pKa = 4.14LTVTVGEE148 pKa = 4.49MPPIAAARR156 pKa = 11.84GEE158 pKa = 4.2VALLSEE164 pKa = 5.36TIPEE168 pKa = 4.12LWVGGVQVTSANASDD183 pKa = 3.92VLGEE187 pKa = 4.05ADD189 pKa = 3.73GDD191 pKa = 4.35GATVIYY197 pKa = 10.41DD198 pKa = 3.83PASGTLTLDD207 pKa = 3.33NANITNGYY215 pKa = 7.46NTTIDD220 pKa = 3.64DD221 pKa = 3.93RR222 pKa = 11.84NVQFGICSTLNALKK236 pKa = 10.63LVVVGTNTITCPNASVKK253 pKa = 10.44DD254 pKa = 3.6SYY256 pKa = 11.68AIYY259 pKa = 8.15TTGTLEE265 pKa = 3.83ISEE268 pKa = 4.58SGSGNLTATGGTASDD283 pKa = 4.01ISAGMYY289 pKa = 10.36VSDD292 pKa = 4.3GLTIEE297 pKa = 4.68SGSLTATGGTAIYY310 pKa = 9.31LQSKK314 pKa = 9.98SYY316 pKa = 10.71GVYY319 pKa = 10.49ADD321 pKa = 3.73SLTIEE326 pKa = 4.76GGSLTANGGAVTYY339 pKa = 10.26QSGEE343 pKa = 4.15SYY345 pKa = 10.74GAYY348 pKa = 10.06ADD350 pKa = 3.84SLTIEE355 pKa = 4.41SGSLIAKK362 pKa = 9.72GGAATGTFGKK372 pKa = 10.41SYY374 pKa = 10.66GVYY377 pKa = 10.04AGNSVSVSNDD387 pKa = 2.6GTLTATGGMASNSGTSAGVYY407 pKa = 10.17LNNGDD412 pKa = 3.89STDD415 pKa = 3.8SNGSVSVSDD424 pKa = 4.13GGILTATGGMASTTSVGVYY443 pKa = 9.92VNNISSNGSTVSVSVSDD460 pKa = 5.1DD461 pKa = 3.8GEE463 pKa = 4.21LTAKK467 pKa = 10.53GGTASGTFGTSVGVYY482 pKa = 9.63FDD484 pKa = 4.42NRR486 pKa = 11.84SSTDD490 pKa = 3.04STGSVSVSGSGKK502 pKa = 8.49LTATGDD508 pKa = 3.65AASGTFGTSAGVYY521 pKa = 9.57FDD523 pKa = 4.53NRR525 pKa = 11.84SSTGSTGSVSVSDD538 pKa = 4.41DD539 pKa = 3.47GKK541 pKa = 9.92LTATGDD547 pKa = 3.77AASGSQGTSAGVYY560 pKa = 9.85VNNNSTDD567 pKa = 3.1STGRR571 pKa = 11.84VSVSGHH577 pKa = 6.19GEE579 pKa = 3.76LTATGGMASATSAGVFSNGSGSSVSVSDD607 pKa = 5.07DD608 pKa = 3.25GSLTATGNAASASYY622 pKa = 10.42GVSAEE627 pKa = 4.13NGSMSIFDD635 pKa = 3.88GTITAQSGAASSTSAGVYY653 pKa = 10.04SGSLSVGGGMVTATGGTASDD673 pKa = 3.7ISGKK677 pKa = 9.97SAGVYY682 pKa = 7.41ATSVSVFGSGEE693 pKa = 3.83LTATGDD699 pKa = 3.68AASEE703 pKa = 4.46TNGTSAGVFSNGSGSSVSVSDD724 pKa = 5.07DD725 pKa = 3.25GSLTATGDD733 pKa = 3.58EE734 pKa = 4.04ASGFVGTSAGVYY746 pKa = 7.81FQSSNGSMTVSGGTVEE762 pKa = 4.28ATGGTASAISGGVYY776 pKa = 9.84FQYY779 pKa = 10.33SGSMTISGGTVTAQSGTAATSQAMNTPPPRR809 pKa = 11.84PP810 pKa = 3.44
MM1 pKa = 7.46NKK3 pKa = 9.88RR4 pKa = 11.84LFSMFLALCMIVTMLPVSAMAEE26 pKa = 4.26EE27 pKa = 4.05IHH29 pKa = 5.41TTIGGSGEE37 pKa = 4.23IISFAPLTEE46 pKa = 3.91TAKK49 pKa = 10.73AVSLGTSIEE58 pKa = 4.06DD59 pKa = 3.61LEE61 pKa = 4.78LPEE64 pKa = 4.5TLTATVRR71 pKa = 11.84TAVPADD77 pKa = 3.72EE78 pKa = 6.49DD79 pKa = 3.99STQDD83 pKa = 3.12SGSPEE88 pKa = 4.04TATPTTAAEE97 pKa = 4.45PKK99 pKa = 9.12WKK101 pKa = 8.4EE102 pKa = 3.99TTGDD106 pKa = 3.89IPVEE110 pKa = 4.1WASPDD115 pKa = 3.27YY116 pKa = 11.79DD117 pKa = 3.45MDD119 pKa = 4.07TEE121 pKa = 4.41GVYY124 pKa = 10.7VFTPVIEE131 pKa = 5.12GYY133 pKa = 8.18TVSAPLPEE141 pKa = 4.14LTVTVGEE148 pKa = 4.49MPPIAAARR156 pKa = 11.84GEE158 pKa = 4.2VALLSEE164 pKa = 5.36TIPEE168 pKa = 4.12LWVGGVQVTSANASDD183 pKa = 3.92VLGEE187 pKa = 4.05ADD189 pKa = 3.73GDD191 pKa = 4.35GATVIYY197 pKa = 10.41DD198 pKa = 3.83PASGTLTLDD207 pKa = 3.33NANITNGYY215 pKa = 7.46NTTIDD220 pKa = 3.64DD221 pKa = 3.93RR222 pKa = 11.84NVQFGICSTLNALKK236 pKa = 10.63LVVVGTNTITCPNASVKK253 pKa = 10.44DD254 pKa = 3.6SYY256 pKa = 11.68AIYY259 pKa = 8.15TTGTLEE265 pKa = 3.83ISEE268 pKa = 4.58SGSGNLTATGGTASDD283 pKa = 4.01ISAGMYY289 pKa = 10.36VSDD292 pKa = 4.3GLTIEE297 pKa = 4.68SGSLTATGGTAIYY310 pKa = 9.31LQSKK314 pKa = 9.98SYY316 pKa = 10.71GVYY319 pKa = 10.49ADD321 pKa = 3.73SLTIEE326 pKa = 4.76GGSLTANGGAVTYY339 pKa = 10.26QSGEE343 pKa = 4.15SYY345 pKa = 10.74GAYY348 pKa = 10.06ADD350 pKa = 3.84SLTIEE355 pKa = 4.41SGSLIAKK362 pKa = 9.72GGAATGTFGKK372 pKa = 10.41SYY374 pKa = 10.66GVYY377 pKa = 10.04AGNSVSVSNDD387 pKa = 2.6GTLTATGGMASNSGTSAGVYY407 pKa = 10.17LNNGDD412 pKa = 3.89STDD415 pKa = 3.8SNGSVSVSDD424 pKa = 4.13GGILTATGGMASTTSVGVYY443 pKa = 9.92VNNISSNGSTVSVSVSDD460 pKa = 5.1DD461 pKa = 3.8GEE463 pKa = 4.21LTAKK467 pKa = 10.53GGTASGTFGTSVGVYY482 pKa = 9.63FDD484 pKa = 4.42NRR486 pKa = 11.84SSTDD490 pKa = 3.04STGSVSVSGSGKK502 pKa = 8.49LTATGDD508 pKa = 3.65AASGTFGTSAGVYY521 pKa = 9.57FDD523 pKa = 4.53NRR525 pKa = 11.84SSTGSTGSVSVSDD538 pKa = 4.41DD539 pKa = 3.47GKK541 pKa = 9.92LTATGDD547 pKa = 3.77AASGSQGTSAGVYY560 pKa = 9.85VNNNSTDD567 pKa = 3.1STGRR571 pKa = 11.84VSVSGHH577 pKa = 6.19GEE579 pKa = 3.76LTATGGMASATSAGVFSNGSGSSVSVSDD607 pKa = 5.07DD608 pKa = 3.25GSLTATGNAASASYY622 pKa = 10.42GVSAEE627 pKa = 4.13NGSMSIFDD635 pKa = 3.88GTITAQSGAASSTSAGVYY653 pKa = 10.04SGSLSVGGGMVTATGGTASDD673 pKa = 3.7ISGKK677 pKa = 9.97SAGVYY682 pKa = 7.41ATSVSVFGSGEE693 pKa = 3.83LTATGDD699 pKa = 3.68AASEE703 pKa = 4.46TNGTSAGVFSNGSGSSVSVSDD724 pKa = 5.07DD725 pKa = 3.25GSLTATGDD733 pKa = 3.58EE734 pKa = 4.04ASGFVGTSAGVYY746 pKa = 7.81FQSSNGSMTVSGGTVEE762 pKa = 4.28ATGGTASAISGGVYY776 pKa = 9.84FQYY779 pKa = 10.33SGSMTISGGTVTAQSGTAATSQAMNTPPPRR809 pKa = 11.84PP810 pKa = 3.44
Molecular weight: 79.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D9R425|D9R425_CLOSW Amino acid carrier protein OS=Clostridium saccharolyticum (strain ATCC 35040 / DSM 2544 / NRCC 2533 / WM1) OX=610130 GN=Closa_0501 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.33GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.33GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 4.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1343575 |
30 |
5128 |
325.1 |
36.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.507 ± 0.04 | 1.39 ± 0.02 |
5.356 ± 0.027 | 7.326 ± 0.046 |
4.15 ± 0.031 | 7.569 ± 0.045 |
1.729 ± 0.017 | 7.299 ± 0.037 |
6.531 ± 0.029 | 9.158 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.167 ± 0.022 | 4.277 ± 0.023 |
3.546 ± 0.03 | 3.227 ± 0.023 |
4.483 ± 0.032 | 6.117 ± 0.033 |
5.415 ± 0.042 | 6.778 ± 0.028 |
0.946 ± 0.014 | 4.029 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
