
Desulfovibrio carbinoliphilus subsp. oakridgensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Desulfovibrio; Desulfovibrio carbinoliphilus
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
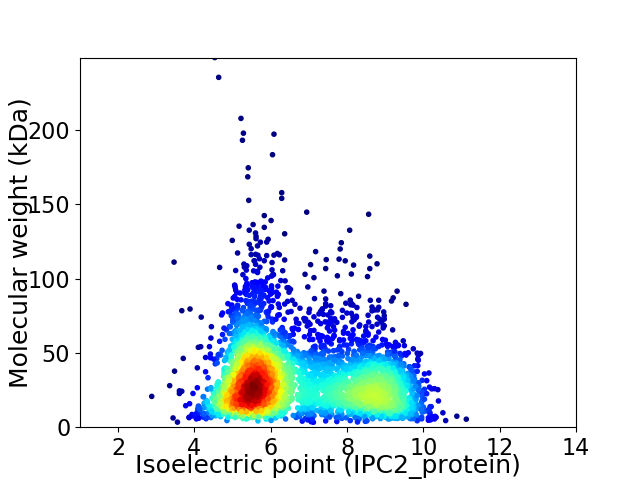
Virtual 2D-PAGE plot for 3713 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G7Q8V4|G7Q8V4_9DELT Uncharacterized protein OS=Desulfovibrio carbinoliphilus subsp. oakridgensis OX=694327 GN=DFW101_1432 PE=4 SV=1
MM1 pKa = 8.02ADD3 pKa = 3.16TGNPGTFLLFSDD15 pKa = 3.53VHH17 pKa = 6.86FDD19 pKa = 3.72PFADD23 pKa = 4.12PSLVPALAASDD34 pKa = 3.7VSAWTGLLSSSSQTAYY50 pKa = 10.43AAYY53 pKa = 9.88GSDD56 pKa = 3.03SNYY59 pKa = 10.4PLFEE63 pKa = 4.36SALDD67 pKa = 3.79DD68 pKa = 3.75MAARR72 pKa = 11.84AADD75 pKa = 3.74VSTILYY81 pKa = 9.46PGDD84 pKa = 3.33ILAHH88 pKa = 6.89DD89 pKa = 5.06FPQDD93 pKa = 3.51YY94 pKa = 10.63AALTGDD100 pKa = 4.05TSQAGLDD107 pKa = 3.39AFAQKK112 pKa = 9.06TVAFFVQEE120 pKa = 3.88VDD122 pKa = 3.77SRR124 pKa = 11.84FPDD127 pKa = 3.49AMVLVADD134 pKa = 5.27GNCDD138 pKa = 3.4TDD140 pKa = 3.71NMSGSIGARR149 pKa = 11.84PGDD152 pKa = 4.12PYY154 pKa = 11.45LSDD157 pKa = 3.69TAPLLAQAFFNNDD170 pKa = 2.82TDD172 pKa = 3.68RR173 pKa = 11.84TAFASTYY180 pKa = 11.02AVGGYY185 pKa = 9.53YY186 pKa = 10.23AVEE189 pKa = 4.22PDD191 pKa = 3.73GPTGLKK197 pKa = 10.61YY198 pKa = 10.26IVLNDD203 pKa = 3.61NLWITEE209 pKa = 3.97YY210 pKa = 11.08DD211 pKa = 3.75DD212 pKa = 4.17PAAGAVEE219 pKa = 4.26LSWFASEE226 pKa = 4.9LAEE229 pKa = 4.91SAQNFQKK236 pKa = 10.5VWVVGHH242 pKa = 6.41IPVGANASSVVASYY256 pKa = 11.12DD257 pKa = 3.16QTGRR261 pKa = 11.84IAYY264 pKa = 9.8SGNLDD269 pKa = 3.88DD270 pKa = 5.45GFNAAFVGLEE280 pKa = 3.92LAYY283 pKa = 10.59DD284 pKa = 3.77ATIKK288 pKa = 10.24ATFTGHH294 pKa = 4.54THH296 pKa = 6.55NDD298 pKa = 3.4DD299 pKa = 3.46FRR301 pKa = 11.84LVTDD305 pKa = 4.54GSGAANLLRR314 pKa = 11.84IAPSISPVFDD324 pKa = 3.94NNPGYY329 pKa = 10.14QVYY332 pKa = 10.7SFDD335 pKa = 3.73TRR337 pKa = 11.84TSSLLDD343 pKa = 3.29EE344 pKa = 4.46TTYY347 pKa = 11.15TLDD350 pKa = 3.88LQSSSPAWSKK360 pKa = 10.51EE361 pKa = 3.5YY362 pKa = 10.56AYY364 pKa = 10.63AEE366 pKa = 4.55TYY368 pKa = 10.15GQTLATPQEE377 pKa = 3.91WQAVYY382 pKa = 10.29GAILTNPATQAAYY395 pKa = 9.42IDD397 pKa = 4.18HH398 pKa = 7.36LGQGAATQFPVTAANLPAYY417 pKa = 9.78LATPGFVLPATYY429 pKa = 10.29NAAVAALAGG438 pKa = 3.81
MM1 pKa = 8.02ADD3 pKa = 3.16TGNPGTFLLFSDD15 pKa = 3.53VHH17 pKa = 6.86FDD19 pKa = 3.72PFADD23 pKa = 4.12PSLVPALAASDD34 pKa = 3.7VSAWTGLLSSSSQTAYY50 pKa = 10.43AAYY53 pKa = 9.88GSDD56 pKa = 3.03SNYY59 pKa = 10.4PLFEE63 pKa = 4.36SALDD67 pKa = 3.79DD68 pKa = 3.75MAARR72 pKa = 11.84AADD75 pKa = 3.74VSTILYY81 pKa = 9.46PGDD84 pKa = 3.33ILAHH88 pKa = 6.89DD89 pKa = 5.06FPQDD93 pKa = 3.51YY94 pKa = 10.63AALTGDD100 pKa = 4.05TSQAGLDD107 pKa = 3.39AFAQKK112 pKa = 9.06TVAFFVQEE120 pKa = 3.88VDD122 pKa = 3.77SRR124 pKa = 11.84FPDD127 pKa = 3.49AMVLVADD134 pKa = 5.27GNCDD138 pKa = 3.4TDD140 pKa = 3.71NMSGSIGARR149 pKa = 11.84PGDD152 pKa = 4.12PYY154 pKa = 11.45LSDD157 pKa = 3.69TAPLLAQAFFNNDD170 pKa = 2.82TDD172 pKa = 3.68RR173 pKa = 11.84TAFASTYY180 pKa = 11.02AVGGYY185 pKa = 9.53YY186 pKa = 10.23AVEE189 pKa = 4.22PDD191 pKa = 3.73GPTGLKK197 pKa = 10.61YY198 pKa = 10.26IVLNDD203 pKa = 3.61NLWITEE209 pKa = 3.97YY210 pKa = 11.08DD211 pKa = 3.75DD212 pKa = 4.17PAAGAVEE219 pKa = 4.26LSWFASEE226 pKa = 4.9LAEE229 pKa = 4.91SAQNFQKK236 pKa = 10.5VWVVGHH242 pKa = 6.41IPVGANASSVVASYY256 pKa = 11.12DD257 pKa = 3.16QTGRR261 pKa = 11.84IAYY264 pKa = 9.8SGNLDD269 pKa = 3.88DD270 pKa = 5.45GFNAAFVGLEE280 pKa = 3.92LAYY283 pKa = 10.59DD284 pKa = 3.77ATIKK288 pKa = 10.24ATFTGHH294 pKa = 4.54THH296 pKa = 6.55NDD298 pKa = 3.4DD299 pKa = 3.46FRR301 pKa = 11.84LVTDD305 pKa = 4.54GSGAANLLRR314 pKa = 11.84IAPSISPVFDD324 pKa = 3.94NNPGYY329 pKa = 10.14QVYY332 pKa = 10.7SFDD335 pKa = 3.73TRR337 pKa = 11.84TSSLLDD343 pKa = 3.29EE344 pKa = 4.46TTYY347 pKa = 11.15TLDD350 pKa = 3.88LQSSSPAWSKK360 pKa = 10.51EE361 pKa = 3.5YY362 pKa = 10.56AYY364 pKa = 10.63AEE366 pKa = 4.55TYY368 pKa = 10.15GQTLATPQEE377 pKa = 3.91WQAVYY382 pKa = 10.29GAILTNPATQAAYY395 pKa = 9.42IDD397 pKa = 4.18HH398 pKa = 7.36LGQGAATQFPVTAANLPAYY417 pKa = 9.78LATPGFVLPATYY429 pKa = 10.29NAAVAALAGG438 pKa = 3.81
Molecular weight: 46.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G7Q6L8|G7Q6L8_9DELT NADH dehydrogenase (Quinone) OS=Desulfovibrio carbinoliphilus subsp. oakridgensis OX=694327 GN=DFW101_1623 PE=4 SV=1
MM1 pKa = 7.43SKK3 pKa = 8.62KK4 pKa = 8.87TYY6 pKa = 9.67QPSKK10 pKa = 9.23IRR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 8.13RR15 pKa = 11.84THH17 pKa = 5.96GFLVRR22 pKa = 11.84SRR24 pKa = 11.84TKK26 pKa = 10.32NGQAILRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.7GRR40 pKa = 11.84KK41 pKa = 8.66RR42 pKa = 11.84LAVV45 pKa = 3.41
MM1 pKa = 7.43SKK3 pKa = 8.62KK4 pKa = 8.87TYY6 pKa = 9.67QPSKK10 pKa = 9.23IRR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 8.13RR15 pKa = 11.84THH17 pKa = 5.96GFLVRR22 pKa = 11.84SRR24 pKa = 11.84TKK26 pKa = 10.32NGQAILRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.7GRR40 pKa = 11.84KK41 pKa = 8.66RR42 pKa = 11.84LAVV45 pKa = 3.41
Molecular weight: 5.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1225608 |
30 |
2352 |
330.1 |
35.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.929 ± 0.076 | 1.281 ± 0.017 |
5.48 ± 0.027 | 5.761 ± 0.041 |
3.872 ± 0.026 | 8.783 ± 0.042 |
1.871 ± 0.017 | 4.223 ± 0.035 |
3.668 ± 0.034 | 11.123 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.327 ± 0.016 | 2.33 ± 0.023 |
5.645 ± 0.039 | 2.746 ± 0.024 |
7.162 ± 0.041 | 4.823 ± 0.03 |
5.032 ± 0.035 | 7.477 ± 0.033 |
1.108 ± 0.015 | 2.359 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
