
Dolphin morbillivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Morbillivirus; Cetacean morbillivirus
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
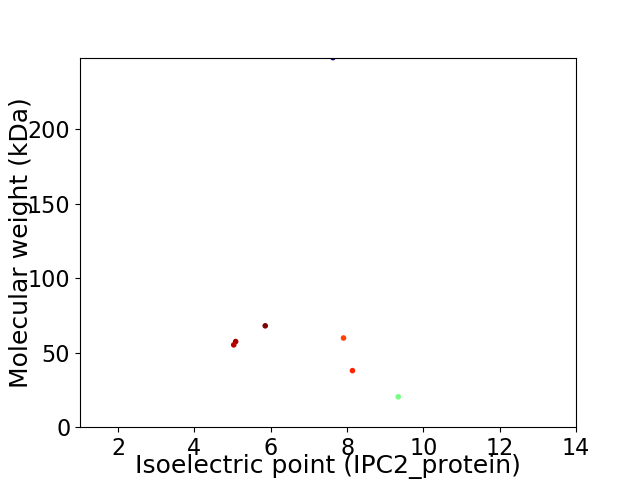
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q709E7|Q709E7_9MONO Isoform of Q709E8 Protein C OS=Dolphin morbillivirus OX=37131 GN=P/V/C PE=3 SV=1
MM1 pKa = 8.2AEE3 pKa = 3.69EE4 pKa = 3.77QAYY7 pKa = 9.6HH8 pKa = 5.39VNKK11 pKa = 10.43GLEE14 pKa = 4.24CLKK17 pKa = 10.54SLRR20 pKa = 11.84EE21 pKa = 4.01NPPDD25 pKa = 3.34AVEE28 pKa = 4.02IKK30 pKa = 9.19EE31 pKa = 4.04AQIIRR36 pKa = 11.84SKK38 pKa = 10.59AACEE42 pKa = 3.9EE43 pKa = 4.48SSEE46 pKa = 4.62SHH48 pKa = 6.35HH49 pKa = 6.42QDD51 pKa = 2.75NSEE54 pKa = 4.0KK55 pKa = 10.26DD56 pKa = 3.33TLDD59 pKa = 3.48FDD61 pKa = 4.91EE62 pKa = 5.34SCSSAIRR69 pKa = 11.84PEE71 pKa = 4.25TYY73 pKa = 10.75RR74 pKa = 11.84MLLGDD79 pKa = 3.81DD80 pKa = 3.56TGFRR84 pKa = 11.84APGYY88 pKa = 9.42IPNEE92 pKa = 4.26GEE94 pKa = 4.5PEE96 pKa = 4.25PGDD99 pKa = 3.31IGKK102 pKa = 9.73EE103 pKa = 4.0EE104 pKa = 3.99PAVRR108 pKa = 11.84CYY110 pKa = 10.36HH111 pKa = 6.32VYY113 pKa = 10.91DD114 pKa = 3.91HH115 pKa = 7.16GGQAVEE121 pKa = 4.43GVKK124 pKa = 10.73DD125 pKa = 3.52ADD127 pKa = 4.03LLVVPTGSDD136 pKa = 3.16DD137 pKa = 3.79DD138 pKa = 4.98AEE140 pKa = 4.27FRR142 pKa = 11.84DD143 pKa = 3.97GDD145 pKa = 3.98EE146 pKa = 4.88SSLEE150 pKa = 3.98SDD152 pKa = 4.12GEE154 pKa = 4.36SGTVDD159 pKa = 2.92TRR161 pKa = 11.84GNSSSNRR168 pKa = 11.84GSAPRR173 pKa = 11.84IKK175 pKa = 10.39VEE177 pKa = 3.84RR178 pKa = 11.84SSDD181 pKa = 3.54VEE183 pKa = 4.48TISSEE188 pKa = 3.96EE189 pKa = 3.78LQGLIRR195 pKa = 11.84SQSQKK200 pKa = 10.79HH201 pKa = 5.53NGFGVDD207 pKa = 3.67RR208 pKa = 11.84FLKK211 pKa = 10.41VPPIPTSVPLDD222 pKa = 3.77PAPKK226 pKa = 9.79SIKK229 pKa = 10.06KK230 pKa = 8.63GTGEE234 pKa = 3.87RR235 pKa = 11.84SALSGTEE242 pKa = 4.17TEE244 pKa = 4.72FSLTGGATRR253 pKa = 11.84LAQEE257 pKa = 4.41SRR259 pKa = 11.84WASSEE264 pKa = 3.81SSAPAEE270 pKa = 4.01NVRR273 pKa = 11.84QSVTNAEE280 pKa = 4.33RR281 pKa = 11.84TQKK284 pKa = 10.2PPQGSGTTASQKK296 pKa = 9.89SQNNGHH302 pKa = 6.43SDD304 pKa = 3.78DD305 pKa = 4.4EE306 pKa = 4.9YY307 pKa = 10.86EE308 pKa = 5.29DD309 pKa = 3.81EE310 pKa = 5.99LFMEE314 pKa = 4.39VQEE317 pKa = 4.41IKK319 pKa = 9.85TAITKK324 pKa = 10.11INEE327 pKa = 4.11DD328 pKa = 3.36NQQIISKK335 pKa = 9.57LDD337 pKa = 3.47SIMLLKK343 pKa = 11.11GEE345 pKa = 4.28IEE347 pKa = 4.43SIKK350 pKa = 10.69KK351 pKa = 9.98QINKK355 pKa = 9.55QNITISTIEE364 pKa = 3.93GHH366 pKa = 6.68LSSIMIAIPGFGKK379 pKa = 10.48DD380 pKa = 4.04PNDD383 pKa = 3.34PTADD387 pKa = 3.42VEE389 pKa = 4.6LNPDD393 pKa = 3.78LRR395 pKa = 11.84PIISRR400 pKa = 11.84DD401 pKa = 3.08AGRR404 pKa = 11.84ALAEE408 pKa = 4.11VLKK411 pKa = 10.81RR412 pKa = 11.84PAVEE416 pKa = 4.31RR417 pKa = 11.84NPKK420 pKa = 8.29VTPKK424 pKa = 10.06VHH426 pKa = 6.85PGSKK430 pKa = 10.19GQILRR435 pKa = 11.84DD436 pKa = 3.79LQLKK440 pKa = 9.54PVDD443 pKa = 3.85RR444 pKa = 11.84KK445 pKa = 9.68MSSAVGFVPTDD456 pKa = 3.69DD457 pKa = 4.04LPSRR461 pKa = 11.84SVLRR465 pKa = 11.84SMIKK469 pKa = 10.12SSNLEE474 pKa = 4.19SEE476 pKa = 5.09HH477 pKa = 6.02KK478 pKa = 10.34RR479 pKa = 11.84SMIGLLNDD487 pKa = 3.4VKK489 pKa = 10.82SGKK492 pKa = 9.91DD493 pKa = 3.06LGEE496 pKa = 4.3FYY498 pKa = 11.4QMVKK502 pKa = 10.53KK503 pKa = 10.62IIKK506 pKa = 9.68
MM1 pKa = 8.2AEE3 pKa = 3.69EE4 pKa = 3.77QAYY7 pKa = 9.6HH8 pKa = 5.39VNKK11 pKa = 10.43GLEE14 pKa = 4.24CLKK17 pKa = 10.54SLRR20 pKa = 11.84EE21 pKa = 4.01NPPDD25 pKa = 3.34AVEE28 pKa = 4.02IKK30 pKa = 9.19EE31 pKa = 4.04AQIIRR36 pKa = 11.84SKK38 pKa = 10.59AACEE42 pKa = 3.9EE43 pKa = 4.48SSEE46 pKa = 4.62SHH48 pKa = 6.35HH49 pKa = 6.42QDD51 pKa = 2.75NSEE54 pKa = 4.0KK55 pKa = 10.26DD56 pKa = 3.33TLDD59 pKa = 3.48FDD61 pKa = 4.91EE62 pKa = 5.34SCSSAIRR69 pKa = 11.84PEE71 pKa = 4.25TYY73 pKa = 10.75RR74 pKa = 11.84MLLGDD79 pKa = 3.81DD80 pKa = 3.56TGFRR84 pKa = 11.84APGYY88 pKa = 9.42IPNEE92 pKa = 4.26GEE94 pKa = 4.5PEE96 pKa = 4.25PGDD99 pKa = 3.31IGKK102 pKa = 9.73EE103 pKa = 4.0EE104 pKa = 3.99PAVRR108 pKa = 11.84CYY110 pKa = 10.36HH111 pKa = 6.32VYY113 pKa = 10.91DD114 pKa = 3.91HH115 pKa = 7.16GGQAVEE121 pKa = 4.43GVKK124 pKa = 10.73DD125 pKa = 3.52ADD127 pKa = 4.03LLVVPTGSDD136 pKa = 3.16DD137 pKa = 3.79DD138 pKa = 4.98AEE140 pKa = 4.27FRR142 pKa = 11.84DD143 pKa = 3.97GDD145 pKa = 3.98EE146 pKa = 4.88SSLEE150 pKa = 3.98SDD152 pKa = 4.12GEE154 pKa = 4.36SGTVDD159 pKa = 2.92TRR161 pKa = 11.84GNSSSNRR168 pKa = 11.84GSAPRR173 pKa = 11.84IKK175 pKa = 10.39VEE177 pKa = 3.84RR178 pKa = 11.84SSDD181 pKa = 3.54VEE183 pKa = 4.48TISSEE188 pKa = 3.96EE189 pKa = 3.78LQGLIRR195 pKa = 11.84SQSQKK200 pKa = 10.79HH201 pKa = 5.53NGFGVDD207 pKa = 3.67RR208 pKa = 11.84FLKK211 pKa = 10.41VPPIPTSVPLDD222 pKa = 3.77PAPKK226 pKa = 9.79SIKK229 pKa = 10.06KK230 pKa = 8.63GTGEE234 pKa = 3.87RR235 pKa = 11.84SALSGTEE242 pKa = 4.17TEE244 pKa = 4.72FSLTGGATRR253 pKa = 11.84LAQEE257 pKa = 4.41SRR259 pKa = 11.84WASSEE264 pKa = 3.81SSAPAEE270 pKa = 4.01NVRR273 pKa = 11.84QSVTNAEE280 pKa = 4.33RR281 pKa = 11.84TQKK284 pKa = 10.2PPQGSGTTASQKK296 pKa = 9.89SQNNGHH302 pKa = 6.43SDD304 pKa = 3.78DD305 pKa = 4.4EE306 pKa = 4.9YY307 pKa = 10.86EE308 pKa = 5.29DD309 pKa = 3.81EE310 pKa = 5.99LFMEE314 pKa = 4.39VQEE317 pKa = 4.41IKK319 pKa = 9.85TAITKK324 pKa = 10.11INEE327 pKa = 4.11DD328 pKa = 3.36NQQIISKK335 pKa = 9.57LDD337 pKa = 3.47SIMLLKK343 pKa = 11.11GEE345 pKa = 4.28IEE347 pKa = 4.43SIKK350 pKa = 10.69KK351 pKa = 9.98QINKK355 pKa = 9.55QNITISTIEE364 pKa = 3.93GHH366 pKa = 6.68LSSIMIAIPGFGKK379 pKa = 10.48DD380 pKa = 4.04PNDD383 pKa = 3.34PTADD387 pKa = 3.42VEE389 pKa = 4.6LNPDD393 pKa = 3.78LRR395 pKa = 11.84PIISRR400 pKa = 11.84DD401 pKa = 3.08AGRR404 pKa = 11.84ALAEE408 pKa = 4.11VLKK411 pKa = 10.81RR412 pKa = 11.84PAVEE416 pKa = 4.31RR417 pKa = 11.84NPKK420 pKa = 8.29VTPKK424 pKa = 10.06VHH426 pKa = 6.85PGSKK430 pKa = 10.19GQILRR435 pKa = 11.84DD436 pKa = 3.79LQLKK440 pKa = 9.54PVDD443 pKa = 3.85RR444 pKa = 11.84KK445 pKa = 9.68MSSAVGFVPTDD456 pKa = 3.69DD457 pKa = 4.04LPSRR461 pKa = 11.84SVLRR465 pKa = 11.84SMIKK469 pKa = 10.12SSNLEE474 pKa = 4.19SEE476 pKa = 5.09HH477 pKa = 6.02KK478 pKa = 10.34RR479 pKa = 11.84SMIGLLNDD487 pKa = 3.4VKK489 pKa = 10.82SGKK492 pKa = 9.91DD493 pKa = 3.06LGEE496 pKa = 4.3FYY498 pKa = 11.4QMVKK502 pKa = 10.53KK503 pKa = 10.62IIKK506 pKa = 9.68
Molecular weight: 55.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q709E7|Q709E7_9MONO Isoform of Q709E8 Protein C OS=Dolphin morbillivirus OX=37131 GN=P/V/C PE=3 SV=1
MM1 pKa = 7.87SIRR4 pKa = 11.84DD5 pKa = 3.67LSVSNLSEE13 pKa = 4.76KK14 pKa = 9.82IRR16 pKa = 11.84PMLSKK21 pKa = 10.39LRR23 pKa = 11.84KK24 pKa = 8.57PKK26 pKa = 10.4LSEE29 pKa = 3.87ARR31 pKa = 11.84PPAKK35 pKa = 10.25NQARR39 pKa = 11.84VITRR43 pKa = 11.84TTPKK47 pKa = 9.19KK48 pKa = 9.13TLLISTNHH56 pKa = 7.04ALQQLDD62 pKa = 3.78QKK64 pKa = 10.22RR65 pKa = 11.84TACYY69 pKa = 10.33LVMIQDD75 pKa = 4.76LEE77 pKa = 4.4HH78 pKa = 6.22QVTSLMKK85 pKa = 10.18EE86 pKa = 4.2SPSQEE91 pKa = 3.54TSEE94 pKa = 4.39RR95 pKa = 11.84RR96 pKa = 11.84NLQYY100 pKa = 11.05DD101 pKa = 3.37VTMFMITAVKK111 pKa = 9.79RR112 pKa = 11.84LKK114 pKa = 9.95EE115 pKa = 4.1SRR117 pKa = 11.84MLTCSWFQQAVMMMQNSEE135 pKa = 4.15TEE137 pKa = 3.8MRR139 pKa = 11.84ALSRR143 pKa = 11.84AMVNLALLIPEE154 pKa = 5.39EE155 pKa = 4.16ILPLTGDD162 pKa = 4.7LLPGLRR168 pKa = 11.84SRR170 pKa = 11.84DD171 pKa = 3.51RR172 pKa = 11.84LTLRR176 pKa = 11.84LL177 pKa = 3.72
MM1 pKa = 7.87SIRR4 pKa = 11.84DD5 pKa = 3.67LSVSNLSEE13 pKa = 4.76KK14 pKa = 9.82IRR16 pKa = 11.84PMLSKK21 pKa = 10.39LRR23 pKa = 11.84KK24 pKa = 8.57PKK26 pKa = 10.4LSEE29 pKa = 3.87ARR31 pKa = 11.84PPAKK35 pKa = 10.25NQARR39 pKa = 11.84VITRR43 pKa = 11.84TTPKK47 pKa = 9.19KK48 pKa = 9.13TLLISTNHH56 pKa = 7.04ALQQLDD62 pKa = 3.78QKK64 pKa = 10.22RR65 pKa = 11.84TACYY69 pKa = 10.33LVMIQDD75 pKa = 4.76LEE77 pKa = 4.4HH78 pKa = 6.22QVTSLMKK85 pKa = 10.18EE86 pKa = 4.2SPSQEE91 pKa = 3.54TSEE94 pKa = 4.39RR95 pKa = 11.84RR96 pKa = 11.84NLQYY100 pKa = 11.05DD101 pKa = 3.37VTMFMITAVKK111 pKa = 9.79RR112 pKa = 11.84LKK114 pKa = 9.95EE115 pKa = 4.1SRR117 pKa = 11.84MLTCSWFQQAVMMMQNSEE135 pKa = 4.15TEE137 pKa = 3.8MRR139 pKa = 11.84ALSRR143 pKa = 11.84AMVNLALLIPEE154 pKa = 5.39EE155 pKa = 4.16ILPLTGDD162 pKa = 4.7LLPGLRR168 pKa = 11.84SRR170 pKa = 11.84DD171 pKa = 3.51RR172 pKa = 11.84LTLRR176 pKa = 11.84LL177 pKa = 3.72
Molecular weight: 20.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4880 |
177 |
2183 |
697.1 |
78.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.799 ± 0.518 | 1.701 ± 0.295 |
5.369 ± 0.515 | 5.84 ± 0.625 |
3.238 ± 0.368 | 6.209 ± 0.419 |
2.152 ± 0.465 | 7.951 ± 0.366 |
5.594 ± 0.257 | 10.369 ± 0.712 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.623 ± 0.271 | 4.488 ± 0.189 |
4.508 ± 0.444 | 3.299 ± 0.31 |
5.758 ± 0.303 | 8.443 ± 0.504 |
5.758 ± 0.212 | 6.25 ± 0.495 |
0.922 ± 0.189 | 3.73 ± 0.634 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
