
Maize streak Reunion virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.52
Get precalculated fractions of proteins
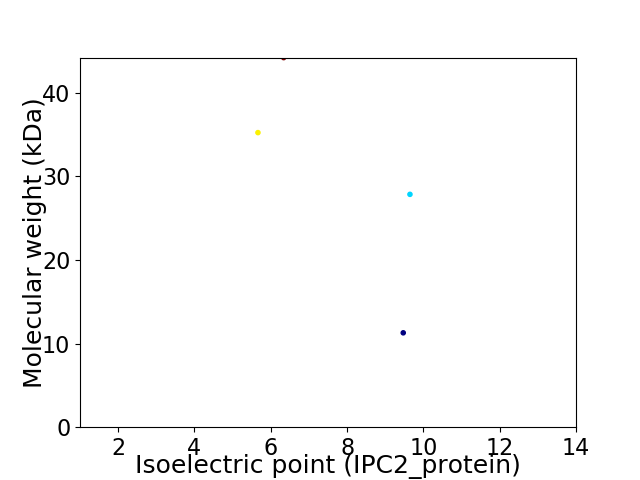
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I1Z771|I1Z771_9GEMI Replication-associated protein OS=Maize streak Reunion virus OX=1182518 PE=3 SV=1
MM1 pKa = 7.37SAFGNHH7 pKa = 5.61FVPTPSVQAEE17 pKa = 4.59VPTPLPGSDD26 pKa = 3.72YY27 pKa = 10.88PSEE30 pKa = 4.16EE31 pKa = 4.78DD32 pKa = 5.77DD33 pKa = 5.24LLQHH37 pKa = 6.33DD38 pKa = 4.88TPVGPDD44 pKa = 3.22PPRR47 pKa = 11.84RR48 pKa = 11.84NFQFKK53 pKa = 10.09SANAFLTYY61 pKa = 9.41PRR63 pKa = 11.84CLLTPFEE70 pKa = 4.97AGQHH74 pKa = 4.6LWEE77 pKa = 4.71VARR80 pKa = 11.84PWTPSYY86 pKa = 10.85ILASSEE92 pKa = 3.95SHH94 pKa = 6.5QDD96 pKa = 3.43GTPHH100 pKa = 6.93LHH102 pKa = 6.71VLLQTIRR109 pKa = 11.84PMSTRR114 pKa = 11.84DD115 pKa = 3.23PGFFDD120 pKa = 3.0IQGYY124 pKa = 8.96HH125 pKa = 6.89PNIQASRR132 pKa = 11.84SPNKK136 pKa = 8.48TRR138 pKa = 11.84EE139 pKa = 4.27YY140 pKa = 10.24ILKK143 pKa = 10.59SPITVYY149 pKa = 10.87SRR151 pKa = 11.84GTFIPRR157 pKa = 11.84AGTSGAGYY165 pKa = 10.62GSTPVPKK172 pKa = 10.03RR173 pKa = 11.84NEE175 pKa = 3.4IMRR178 pKa = 11.84GIIEE182 pKa = 4.49TTTSKK187 pKa = 11.24AEE189 pKa = 4.04YY190 pKa = 9.88LSEE193 pKa = 4.13VQKK196 pKa = 11.14AFPFEE201 pKa = 4.21WATKK205 pKa = 9.04LQQFEE210 pKa = 4.58YY211 pKa = 10.03SAEE214 pKa = 4.05RR215 pKa = 11.84LFPTLPSPFVPPHH228 pKa = 6.66PPSEE232 pKa = 4.59PDD234 pKa = 3.13LTCYY238 pKa = 8.01EE239 pKa = 4.94TIRR242 pKa = 11.84SWKK245 pKa = 10.08DD246 pKa = 3.03EE247 pKa = 4.47NIFQVHH253 pKa = 6.67PYY255 pKa = 10.07IYY257 pKa = 9.4MLEE260 pKa = 4.45HH261 pKa = 6.46PHH263 pKa = 5.99CQSVEE268 pKa = 3.89QAAEE272 pKa = 3.93SLKK275 pKa = 10.9WMDD278 pKa = 2.93EE279 pKa = 4.27CTRR282 pKa = 11.84GMLLHH287 pKa = 7.18APQDD291 pKa = 3.81QGASTSLGLHH301 pKa = 5.45ARR303 pKa = 11.84ARR305 pKa = 11.84LPGPGASTQQ314 pKa = 3.23
MM1 pKa = 7.37SAFGNHH7 pKa = 5.61FVPTPSVQAEE17 pKa = 4.59VPTPLPGSDD26 pKa = 3.72YY27 pKa = 10.88PSEE30 pKa = 4.16EE31 pKa = 4.78DD32 pKa = 5.77DD33 pKa = 5.24LLQHH37 pKa = 6.33DD38 pKa = 4.88TPVGPDD44 pKa = 3.22PPRR47 pKa = 11.84RR48 pKa = 11.84NFQFKK53 pKa = 10.09SANAFLTYY61 pKa = 9.41PRR63 pKa = 11.84CLLTPFEE70 pKa = 4.97AGQHH74 pKa = 4.6LWEE77 pKa = 4.71VARR80 pKa = 11.84PWTPSYY86 pKa = 10.85ILASSEE92 pKa = 3.95SHH94 pKa = 6.5QDD96 pKa = 3.43GTPHH100 pKa = 6.93LHH102 pKa = 6.71VLLQTIRR109 pKa = 11.84PMSTRR114 pKa = 11.84DD115 pKa = 3.23PGFFDD120 pKa = 3.0IQGYY124 pKa = 8.96HH125 pKa = 6.89PNIQASRR132 pKa = 11.84SPNKK136 pKa = 8.48TRR138 pKa = 11.84EE139 pKa = 4.27YY140 pKa = 10.24ILKK143 pKa = 10.59SPITVYY149 pKa = 10.87SRR151 pKa = 11.84GTFIPRR157 pKa = 11.84AGTSGAGYY165 pKa = 10.62GSTPVPKK172 pKa = 10.03RR173 pKa = 11.84NEE175 pKa = 3.4IMRR178 pKa = 11.84GIIEE182 pKa = 4.49TTTSKK187 pKa = 11.24AEE189 pKa = 4.04YY190 pKa = 9.88LSEE193 pKa = 4.13VQKK196 pKa = 11.14AFPFEE201 pKa = 4.21WATKK205 pKa = 9.04LQQFEE210 pKa = 4.58YY211 pKa = 10.03SAEE214 pKa = 4.05RR215 pKa = 11.84LFPTLPSPFVPPHH228 pKa = 6.66PPSEE232 pKa = 4.59PDD234 pKa = 3.13LTCYY238 pKa = 8.01EE239 pKa = 4.94TIRR242 pKa = 11.84SWKK245 pKa = 10.08DD246 pKa = 3.03EE247 pKa = 4.47NIFQVHH253 pKa = 6.67PYY255 pKa = 10.07IYY257 pKa = 9.4MLEE260 pKa = 4.45HH261 pKa = 6.46PHH263 pKa = 5.99CQSVEE268 pKa = 3.89QAAEE272 pKa = 3.93SLKK275 pKa = 10.9WMDD278 pKa = 2.93EE279 pKa = 4.27CTRR282 pKa = 11.84GMLLHH287 pKa = 7.18APQDD291 pKa = 3.81QGASTSLGLHH301 pKa = 5.45ARR303 pKa = 11.84ARR305 pKa = 11.84LPGPGASTQQ314 pKa = 3.23
Molecular weight: 35.24 kDa
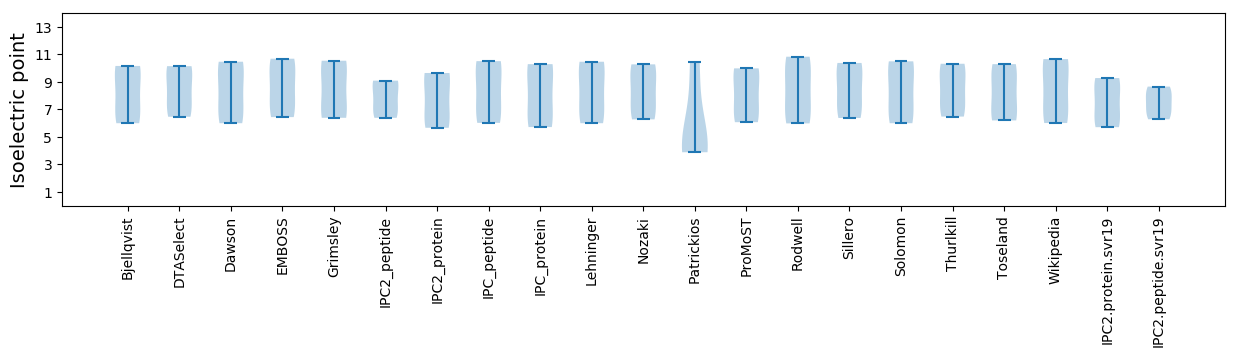
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I1Z770|I1Z770_9GEMI Replication-associated protein OS=Maize streak Reunion virus OX=1182518 PE=3 SV=1
MM1 pKa = 7.54AGSSRR6 pKa = 11.84KK7 pKa = 7.21RR8 pKa = 11.84TRR10 pKa = 11.84RR11 pKa = 11.84GDD13 pKa = 3.44EE14 pKa = 3.96VAWNSKK20 pKa = 8.71RR21 pKa = 11.84SRR23 pKa = 11.84LSAQRR28 pKa = 11.84NPSKK32 pKa = 10.42RR33 pKa = 11.84VPRR36 pKa = 11.84NPRR39 pKa = 11.84VRR41 pKa = 11.84PSLQIQTITNGSSSMVPVKK60 pKa = 10.74QPGYY64 pKa = 10.02CGLLGTYY71 pKa = 10.13SRR73 pKa = 11.84GSDD76 pKa = 2.94EE77 pKa = 4.39NQRR80 pKa = 11.84HH81 pKa = 4.76TSEE84 pKa = 3.83TMTYY88 pKa = 10.29KK89 pKa = 10.36IALDD93 pKa = 3.36LHH95 pKa = 6.37FSITSAAAAYY105 pKa = 9.39SNSGTGVLWLIYY117 pKa = 10.09DD118 pKa = 3.92SQPNGAQPTLKK129 pKa = 10.68DD130 pKa = 2.84IFAYY134 pKa = 10.07EE135 pKa = 4.31DD136 pKa = 3.88SLVAWPYY143 pKa = 6.66TWKK146 pKa = 10.46VSRR149 pKa = 11.84EE150 pKa = 3.81VCHH153 pKa = 6.41RR154 pKa = 11.84FVVKK158 pKa = 10.56RR159 pKa = 11.84RR160 pKa = 11.84YY161 pKa = 8.55TFTLEE166 pKa = 4.1SNGRR170 pKa = 11.84RR171 pKa = 11.84NDD173 pKa = 3.37EE174 pKa = 4.06QPPSNSVWPPCKK186 pKa = 8.79THH188 pKa = 7.51LYY190 pKa = 8.75FHH192 pKa = 7.24KK193 pKa = 9.99FAKK196 pKa = 10.28GLGVRR201 pKa = 11.84TEE203 pKa = 4.13WKK205 pKa = 9.99NDD207 pKa = 3.26TAGSVGNIKK216 pKa = 10.23KK217 pKa = 9.72GALYY221 pKa = 10.24IGIAPGNGVEE231 pKa = 4.44FNVFGKK237 pKa = 8.51TRR239 pKa = 11.84LYY241 pKa = 10.33FKK243 pKa = 10.94SIGNQQ248 pKa = 3.0
MM1 pKa = 7.54AGSSRR6 pKa = 11.84KK7 pKa = 7.21RR8 pKa = 11.84TRR10 pKa = 11.84RR11 pKa = 11.84GDD13 pKa = 3.44EE14 pKa = 3.96VAWNSKK20 pKa = 8.71RR21 pKa = 11.84SRR23 pKa = 11.84LSAQRR28 pKa = 11.84NPSKK32 pKa = 10.42RR33 pKa = 11.84VPRR36 pKa = 11.84NPRR39 pKa = 11.84VRR41 pKa = 11.84PSLQIQTITNGSSSMVPVKK60 pKa = 10.74QPGYY64 pKa = 10.02CGLLGTYY71 pKa = 10.13SRR73 pKa = 11.84GSDD76 pKa = 2.94EE77 pKa = 4.39NQRR80 pKa = 11.84HH81 pKa = 4.76TSEE84 pKa = 3.83TMTYY88 pKa = 10.29KK89 pKa = 10.36IALDD93 pKa = 3.36LHH95 pKa = 6.37FSITSAAAAYY105 pKa = 9.39SNSGTGVLWLIYY117 pKa = 10.09DD118 pKa = 3.92SQPNGAQPTLKK129 pKa = 10.68DD130 pKa = 2.84IFAYY134 pKa = 10.07EE135 pKa = 4.31DD136 pKa = 3.88SLVAWPYY143 pKa = 6.66TWKK146 pKa = 10.46VSRR149 pKa = 11.84EE150 pKa = 3.81VCHH153 pKa = 6.41RR154 pKa = 11.84FVVKK158 pKa = 10.56RR159 pKa = 11.84RR160 pKa = 11.84YY161 pKa = 8.55TFTLEE166 pKa = 4.1SNGRR170 pKa = 11.84RR171 pKa = 11.84NDD173 pKa = 3.37EE174 pKa = 4.06QPPSNSVWPPCKK186 pKa = 8.79THH188 pKa = 7.51LYY190 pKa = 8.75FHH192 pKa = 7.24KK193 pKa = 9.99FAKK196 pKa = 10.28GLGVRR201 pKa = 11.84TEE203 pKa = 4.13WKK205 pKa = 9.99NDD207 pKa = 3.26TAGSVGNIKK216 pKa = 10.23KK217 pKa = 9.72GALYY221 pKa = 10.24IGIAPGNGVEE231 pKa = 4.44FNVFGKK237 pKa = 8.51TRR239 pKa = 11.84LYY241 pKa = 10.33FKK243 pKa = 10.94SIGNQQ248 pKa = 3.0
Molecular weight: 27.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1051 |
102 |
387 |
262.8 |
29.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.994 ± 0.421 | 1.237 ± 0.047 |
4.091 ± 0.383 | 5.138 ± 0.799 |
4.662 ± 0.322 | 6.565 ± 0.771 |
2.759 ± 0.538 | 4.662 ± 0.525 |
4.186 ± 0.934 | 6.946 ± 0.499 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.522 ± 0.165 | 3.52 ± 0.895 |
9.99 ± 1.329 | 4.567 ± 0.384 |
6.565 ± 0.762 | 8.088 ± 0.759 |
7.326 ± 0.313 | 5.614 ± 0.853 |
1.998 ± 0.175 | 4.567 ± 0.427 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
