
Lacinutrix sp. Bg11-31
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Lacinutrix; unclassified Lacinutrix
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
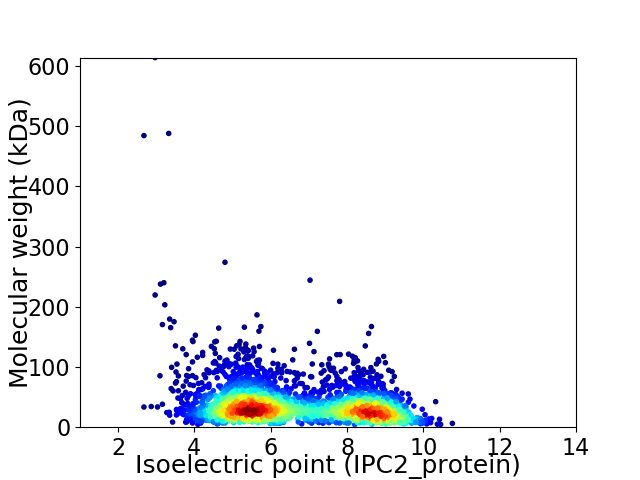
Virtual 2D-PAGE plot for 3170 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K8XII7|A0A2K8XII7_9FLAO Ferredoxin OS=Lacinutrix sp. Bg11-31 OX=2057808 GN=CW733_10870 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 10.19KK3 pKa = 10.08QLILGLAILASVFSCTTDD21 pKa = 3.26DD22 pKa = 3.83TSDD25 pKa = 2.93IVINSTTNNTTGGGGEE41 pKa = 4.37TDD43 pKa = 3.36PATIFLSGTYY53 pKa = 9.67TEE55 pKa = 5.9DD56 pKa = 3.47LTLDD60 pKa = 3.42ANNTYY65 pKa = 10.12KK66 pKa = 10.62INGSLIMASGTTLTIPPCMTIEE88 pKa = 4.24ALSSGADD95 pKa = 3.24VYY97 pKa = 11.31VAISQGAKK105 pKa = 9.78IIANGTASCPIVFTSDD121 pKa = 2.8SSNPLAGDD129 pKa = 3.14WGGLILLGKK138 pKa = 10.5APINSVTGTATATSEE153 pKa = 4.27IASLPYY159 pKa = 10.48GGNTANDD166 pKa = 3.43NSGSLSYY173 pKa = 11.21VRR175 pKa = 11.84VEE177 pKa = 4.31YY178 pKa = 10.96SGGAADD184 pKa = 4.97GQSEE188 pKa = 4.47NNGFSFYY195 pKa = 11.24GVGNGTTVNHH205 pKa = 5.88IQAIEE210 pKa = 4.09GKK212 pKa = 9.99DD213 pKa = 3.29DD214 pKa = 4.46GIEE217 pKa = 4.17FFGGTVNASFISVINAEE234 pKa = 4.64DD235 pKa = 4.63DD236 pKa = 3.96SVDD239 pKa = 3.02WTEE242 pKa = 4.47GFSGTLTDD250 pKa = 3.68VYY252 pKa = 10.61ISNRR256 pKa = 11.84ATDD259 pKa = 3.86DD260 pKa = 3.4KK261 pKa = 11.4AIEE264 pKa = 4.09ADD266 pKa = 4.02GYY268 pKa = 9.64NTDD271 pKa = 4.01FSNATGVFSKK281 pKa = 9.21PTLNNVTIVGEE292 pKa = 4.49GSANSSEE299 pKa = 4.09AVRR302 pKa = 11.84LRR304 pKa = 11.84AGTQGIFSNIHH315 pKa = 4.22ITGYY319 pKa = 11.37AEE321 pKa = 5.66GFDD324 pKa = 5.84LDD326 pKa = 5.98DD327 pKa = 5.72LDD329 pKa = 4.09TGNGVVSDD337 pKa = 4.38DD338 pKa = 3.99LQVTGVTFVDD348 pKa = 3.61VTLNMKK354 pKa = 10.4NDD356 pKa = 3.53TTVTFTIADD365 pKa = 4.89FYY367 pKa = 10.28TNEE370 pKa = 3.88GTATGTDD377 pKa = 3.68YY378 pKa = 7.61TTWGANWTVQQ388 pKa = 3.27
MM1 pKa = 7.67KK2 pKa = 10.19KK3 pKa = 10.08QLILGLAILASVFSCTTDD21 pKa = 3.26DD22 pKa = 3.83TSDD25 pKa = 2.93IVINSTTNNTTGGGGEE41 pKa = 4.37TDD43 pKa = 3.36PATIFLSGTYY53 pKa = 9.67TEE55 pKa = 5.9DD56 pKa = 3.47LTLDD60 pKa = 3.42ANNTYY65 pKa = 10.12KK66 pKa = 10.62INGSLIMASGTTLTIPPCMTIEE88 pKa = 4.24ALSSGADD95 pKa = 3.24VYY97 pKa = 11.31VAISQGAKK105 pKa = 9.78IIANGTASCPIVFTSDD121 pKa = 2.8SSNPLAGDD129 pKa = 3.14WGGLILLGKK138 pKa = 10.5APINSVTGTATATSEE153 pKa = 4.27IASLPYY159 pKa = 10.48GGNTANDD166 pKa = 3.43NSGSLSYY173 pKa = 11.21VRR175 pKa = 11.84VEE177 pKa = 4.31YY178 pKa = 10.96SGGAADD184 pKa = 4.97GQSEE188 pKa = 4.47NNGFSFYY195 pKa = 11.24GVGNGTTVNHH205 pKa = 5.88IQAIEE210 pKa = 4.09GKK212 pKa = 9.99DD213 pKa = 3.29DD214 pKa = 4.46GIEE217 pKa = 4.17FFGGTVNASFISVINAEE234 pKa = 4.64DD235 pKa = 4.63DD236 pKa = 3.96SVDD239 pKa = 3.02WTEE242 pKa = 4.47GFSGTLTDD250 pKa = 3.68VYY252 pKa = 10.61ISNRR256 pKa = 11.84ATDD259 pKa = 3.86DD260 pKa = 3.4KK261 pKa = 11.4AIEE264 pKa = 4.09ADD266 pKa = 4.02GYY268 pKa = 9.64NTDD271 pKa = 4.01FSNATGVFSKK281 pKa = 9.21PTLNNVTIVGEE292 pKa = 4.49GSANSSEE299 pKa = 4.09AVRR302 pKa = 11.84LRR304 pKa = 11.84AGTQGIFSNIHH315 pKa = 4.22ITGYY319 pKa = 11.37AEE321 pKa = 5.66GFDD324 pKa = 5.84LDD326 pKa = 5.98DD327 pKa = 5.72LDD329 pKa = 4.09TGNGVVSDD337 pKa = 4.38DD338 pKa = 3.99LQVTGVTFVDD348 pKa = 3.61VTLNMKK354 pKa = 10.4NDD356 pKa = 3.53TTVTFTIADD365 pKa = 4.89FYY367 pKa = 10.28TNEE370 pKa = 3.88GTATGTDD377 pKa = 3.68YY378 pKa = 7.61TTWGANWTVQQ388 pKa = 3.27
Molecular weight: 40.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K8XDL2|A0A2K8XDL2_9FLAO Uncharacterized protein OS=Lacinutrix sp. Bg11-31 OX=2057808 GN=CW733_02270 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
Molecular weight: 6.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1077573 |
37 |
5836 |
339.9 |
38.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.433 ± 0.048 | 0.784 ± 0.017 |
5.686 ± 0.059 | 6.446 ± 0.047 |
5.242 ± 0.045 | 6.172 ± 0.046 |
1.648 ± 0.024 | 8.178 ± 0.041 |
8.068 ± 0.084 | 9.201 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.032 ± 0.026 | 6.679 ± 0.058 |
3.124 ± 0.026 | 3.205 ± 0.025 |
3.026 ± 0.037 | 6.518 ± 0.034 |
6.395 ± 0.109 | 6.222 ± 0.042 |
0.949 ± 0.015 | 3.993 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
